Naïve Huntington's disease microglia mount a normal response to inflammatory stimuli but display a partially impaired development of innate immune tolerance that can be counteracted by ganglioside GM1
- PMID: 37996924
- PMCID: PMC10668379
- DOI: 10.1186/s12974-023-02963-y
Naïve Huntington's disease microglia mount a normal response to inflammatory stimuli but display a partially impaired development of innate immune tolerance that can be counteracted by ganglioside GM1
Abstract
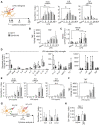
Chronic activation and dysfunction of microglia have been implicated in the pathogenesis and progression of many neurodegenerative disorders, including Huntington's disease (HD). HD is a genetic condition caused by a mutation that affects the folding and function of huntingtin (HTT). Signs of microglia activation have been observed in HD patients even before the onset of symptoms. It is unclear, however, whether pro-inflammatory microglia activation in HD results from cell-autonomous expression of mutant HTT, is the response of microglia to a diseased brain environment, or both. In this study, we used primary microglia isolated from HD knock-in (Q140) and wild-type (Q7) mice to investigate their response to inflammatory conditions in vitro in the absence of confounding effects arising from brain pathology. We show that naïve Q140 microglia do not undergo spontaneous pro-inflammatory activation and respond to inflammatory triggers, including stimulation of TLR4 and TLR2 and exposure to necrotic cells, with similar kinetics of pro-inflammatory gene expression as wild-type microglia. Upon termination of the inflammatory insult, the transcription of pro-inflammatory cytokines is tapered off in Q140 and wild-type microglia with similar kinetics. However, the ability of Q140 microglia to develop tolerance in response to repeated inflammatory stimulations is partially impaired in vitro and in vivo, potentially contributing to the establishment of chronic neuroinflammation in HD. We further show that ganglioside GM1, a glycosphingolipid with anti-inflammatory effects on wild-type microglia, not only decreases the production of pro-inflammatory cytokines and nitric oxide in activated Q140 microglia, but also dramatically dampen microglia response to re-stimulation with LPS in an experimental model of tolerance. These effects are independent from the expression of interleukin 1 receptor associated kinase 3 (Irak-3), a strong modulator of LPS signaling involved in the development of innate immune tolerance and previously shown to be upregulated by immune cell treatment with gangliosides. Altogether, our data suggest that external triggers are required for HD microglia activation, but a cell-autonomous dysfunction that affects the ability of HD microglia to acquire tolerance might contribute to the establishment of neuroinflammation in HD. Administration of GM1 might be beneficial to attenuate chronic microglia activation and neuroinflammation.
Keywords: GM1; Ganglioside; Huntington’s disease; LPS; Microglia; Neuroinflammation; Q140/140 knock-in mice; TLR2; TLR4; Tolerance.
© 2023. The Author(s).
Conflict of interest statement
SS and the University of Alberta hold a patent for the use of GM1 in HD. There are no other competing interests to declare.
Figures

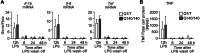




Similar articles
-
Anti-inflammatory role of GM1 and other gangliosides on microglia.J Neuroinflammation. 2022 Jan 6;19(1):9. doi: 10.1186/s12974-021-02374-x. J Neuroinflammation. 2022. PMID: 34991625 Free PMC article.
-
Disease-modifying effects of ganglioside GM1 in Huntington's disease models.EMBO Mol Med. 2017 Nov;9(11):1537-1557. doi: 10.15252/emmm.201707763. EMBO Mol Med. 2017. PMID: 28993428 Free PMC article.
-
Impaired ganglioside metabolism in Huntington's disease and neuroprotective role of GM1.J Neurosci. 2010 Mar 17;30(11):4072-80. doi: 10.1523/JNEUROSCI.6348-09.2010. J Neurosci. 2010. PMID: 20237277 Free PMC article.
-
Neuroinflammation in Huntington's Disease: A Starring Role for Astrocyte and Microglia.Curr Neuropharmacol. 2022;20(6):1116-1143. doi: 10.2174/1570159X19666211201094608. Curr Neuropharmacol. 2022. PMID: 34852742 Free PMC article. Review.
-
The choreography of neuroinflammation in Huntington's disease.Trends Immunol. 2015 Jun;36(6):364-73. doi: 10.1016/j.it.2015.04.007. Epub 2015 May 20. Trends Immunol. 2015. PMID: 26001312 Free PMC article. Review.
Cited by
-
Metabolic Endotoxemia: From the Gut to Neurodegeneration.Int J Mol Sci. 2024 Jun 26;25(13):7006. doi: 10.3390/ijms25137006. Int J Mol Sci. 2024. PMID: 39000116 Free PMC article. Review.
References
-
- A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72(6):971–83. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

