Transcriptomic landscape based on annotated clinical features reveals PLPP2 involvement in lipid raft-mediated proliferation signature of early-stage lung adenocarcinoma
- PMID: 37996944
- PMCID: PMC10666437
- DOI: 10.1186/s13046-023-02877-w
Transcriptomic landscape based on annotated clinical features reveals PLPP2 involvement in lipid raft-mediated proliferation signature of early-stage lung adenocarcinoma
Abstract
Background: Image-based screening improves the detection of early-stage lung adenocarcinoma (LUAD)but also highlights the issue of high false-positive diagnoses, which puts patients at a risk of unnecessary over-treatment. Therefore, more precise discrimination criteria are required to ensure that patients with early-stage LUAD receive appropriate treatments.
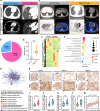
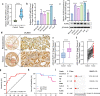
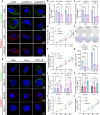
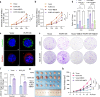
Methods: We integrated 158 early-stage LUAD cases from 2 independent cohorts, including 30 matched resected specimens with complete radiological and pathological information, and 128 retrospective pathological pair-samples with partial follow-up data. This integration allowed us to conduct a correlation analysis between clinical phenotype and transcriptome landscape. Immunohistochemistry was performed using tissue microarrays to examine the expression of phospholipid phosphatase 2 (PLPP2) and lipid-raft markers. Lipidomics analysis was used to determine the changes of lipid components in PLPP2-overexpressed cells. To assess the effects of PLPP2 on the malignant phenotypes of LUAD cells, we conducted mice tumor-bearing experiments and in vitro cellular experiments by knocking down PLPP2 and inhibiting lipid raft synthesis with MβCD, respectively.
Results: Bioinformatics analysis indicated that the co-occurrence of lipid raft formation and rapid cell proliferation might exhibit synergistic effects in driving oncogenesis from lung preneoplasia to adenocarcinoma. The enhanced activation of the cell cycle promoted the transition from non-invasive to invasive status in early-stage LUAD, which was related to an increase in lipid rafts within LUAD cells. PLPP2 participated in lipid raft formation by altering the component contents of lipid rafts, such as esters, sphingomyelin, and sphingosine. Furthermore, elevated PLPP2 levels were identified as an independent prognostic risk factor for LUAD patients. Further results from in vivo and in vitro experiments confirmed that PLPP2 could induce excessive cell proliferation by enhancing lipid raft formation in LUAD cells.
Conclusions: Our study has revealed the characteristics of gene expression profiles in early-stage LUAD patients with the different radiological and pathological subtypes, as well as deciphered transcriptomic evolution trajectory from preneoplasia to invasive LUAD. Furthermore, it suggests that PLPP2-mediated lipid raft synthesis may be a significant biological event in the initiation of early-stage LUAD, offering a potential target for more precise diagnosis and therapy in clinical settings.
Keywords: Cell proliferation; Early stage lung adenocarcinoma; Lipid rafts; Phospholipid phosphatase 2; Transcriptome sequencing.
© 2023. The Author(s).
Conflict of interest statement
The authors have no conflicts of interests and declare no competing financial interests.
Figures



References
-
- Tsutani Y, Miyata Y, Nakayama H, Okumura S, Adachi S, Yoshimura M, et al. Oncologic outcomes of segmentectomy compared with lobectomy for clinical stage IA lung adenocarcinoma: propensity score-matched analysis in a multicenter study. J Thorac Cardiovasc Surg. 2013;146(2):358–364. doi: 10.1016/j.jtcvs.2013.02.008. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- No. 82073242/National Natural Science Foundation of China
- No. 82103421/National Natural Science Foundation of China
- No. 1655187410844/Science and Technology Foundation of Liaoning Province
- NO.2021JH2/10300088/Science and Technology Foundation of Liaoning Province
- NO.30B/Shengjing Hospital of China Medical University 345 Talent Project
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

