Model validation and selection in metabolic flux analysis and flux balance analysis
- PMID: 37997613
- PMCID: PMC10922127
- DOI: 10.1002/btpr.3413
Model validation and selection in metabolic flux analysis and flux balance analysis
Abstract
13C-Metabolic Flux Analysis (13C-MFA) and Flux Balance Analysis (FBA) are widely used to investigate the operation of biochemical networks in both biological and biotechnological research. Both methods use metabolic reaction network models of metabolism operating at steady state so that reaction rates (fluxes) and the levels of metabolic intermediates are constrained to be invariant. They provide estimated (MFA) or predicted (FBA) values of the fluxes through the network in vivo, which cannot be measured directly. These fluxes can shed light on basic biology and have been successfully used to inform metabolic engineering strategies. Several approaches have been taken to test the reliability of estimates and predictions from constraint-based methods and to compare alternative model architectures. Despite advances in other areas of the statistical evaluation of metabolic models, such as the quantification of flux estimate uncertainty, validation and model selection methods have been underappreciated and underexplored. We review the history and state-of-the-art in constraint-based metabolic model validation and model selection. Applications and limitations of the χ2 -test of goodness-of-fit, the most widely used quantitative validation and selection approach in 13C-MFA, are discussed, and complementary and alternative forms of validation and selection are proposed. A combined model validation and selection framework for 13C-MFA incorporating metabolite pool size information that leverages new developments in the field is presented and advocated for. Finally, we discuss how adopting robust validation and selection procedures can enhance confidence in constraint-based modeling as a whole and ultimately facilitate more widespread use of FBA in biotechnology.
Keywords: constraint-based modeling; flux balance analysis; metabolic flux analysis; metabolic modeling; model selection; model validation.
© 2023 The Authors. Biotechnology Progress published by Wiley Periodicals LLC on behalf of American Institute of Chemical Engineers.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures
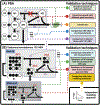

Update of
-
Model Validation and Selection in Metabolic Flux Analysis and Flux Balance Analysis.ArXiv [Preprint]. 2023 Mar 22:arXiv:2303.12651v1. ArXiv. 2023. Update in: Biotechnol Prog. 2024 Jan-Feb;40(1):e3413. doi: 10.1002/btpr.3413. PMID: 36994165 Free PMC article. Updated. Preprint.
References
-
- Koffas MAG, Jung GY, Stephanopoulos G. Engineering metabolism and product formation in Corynebacterium glutamicum by coordinated gene overexpression. Metabolic Engineering. 2003;5(1):32–41. - PubMed
-
- Koffas MAG, Stephanopoulos G. Strain improvement by metabolic engineering: Lysine production as a case study for systems biology. Current Opinion in Biotechnology. 2005;16(3 SPEC. ISS.):361–6. - PubMed
-
- Becker J, Zelder O, Häfner S, Schröder H, Wittmann C. From zero to hero-Design-based systems metabolic engineering of Corynebacterium glutamicum for l-lysine production. Metabolic Engineering. 2011;13(2):159–68. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

