Four New Endophytic Apiospora Species Isolated from Three Dicranopteris Species in Guizhou, China
- PMID: 37998901
- PMCID: PMC10672413
- DOI: 10.3390/jof9111096
Four New Endophytic Apiospora Species Isolated from Three Dicranopteris Species in Guizhou, China
Abstract
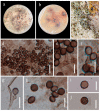
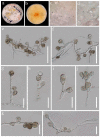
Endophytic fungi isolated from medicinal ferns serve as significant natural resources for drug precursors or bioactive metabolites. During our survey on the diversity of endophytic fungi from Dicranopteris species (a genus of medicinal ferns) in Guizhou, Apoiospora was observed as a dominant fungal group. In this study, seven Apiospora strains, representing four new species, were obtained from the healthy plant tissues of three Dicranopteris species-D. ampla, D. linearis, and D. pedata. The four new species, namely Apiospora aseptata, A. dematiacea, A. dicranopteridis, and A. globosa, were described in detail with color photographs and subjected to phylogenetic analyses using combined LSU, ITS, TEF1-α, and TUB2 sequence data. This study also documented three new hosts for Apiospora species.
Keywords: asexual morph; dematiaceous conidia; medicinal ferns; phylogeny; seven taxa; taxonomy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Hyde K., Fröhlich J., Taylor J. Fungi from palms. XXXVI. Reflections on unitunicate ascomycetes with apiospores. Sydowia. 1998;50:21–80.
-
- Senanayake I.C., Maharachchikumbura S.S.N., Hyde K.D., Bhat J.D., Jones E.G., McKenzie E.H., Dai D.Q., Daranagama D.A., Dayarathne M.C., Goonasekara I.D. Towards unraveling relationships in Xylariomycetidae (Sordariomycetes) Fungal Divers. 2015;73:73–144. doi: 10.1007/s13225-015-0340-y. - DOI
-
- Hyde K.D., Norphanphoun C., Maharachchikumbura S., Bhat D., Jones E., Bundhun D., Chen Y., Bao D., Boonmee S., Calabon M. Refined families of Sordariomycetes. Mycosphere. 2020;11:305–1059. doi: 10.5943/mycosphere/11/1/7. - DOI
-
- Samarakoon M.C., Hyde K.D., Maharachchikumbura S.S., Stadler M., Gareth Jones E., Promputtha I., Suwannarach N., Camporesi E., Bulgakov T.S., Liu J.-K. Taxonomy, phylogeny, molecular dating and ancestral state reconstruction of Xylariomycetidae (Sordariomycetes) Fungal Divers. 2022;112:1–88. doi: 10.1007/s13225-021-00495-5. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

