Proteomics of Paracoccidioides lutzii: Overview of Changes Triggered by Nitrogen Catabolite Repression
- PMID: 37998907
- PMCID: PMC10672198
- DOI: 10.3390/jof9111102
Proteomics of Paracoccidioides lutzii: Overview of Changes Triggered by Nitrogen Catabolite Repression
Abstract
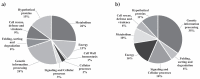
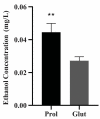
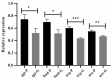
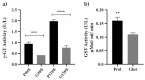
Members of the Paracoccidioides complex are the causative agents of Paracoccidioidomycosis (PCM), a human systemic mycosis endemic in Latin America. Upon initial contact with the host, the pathogen needs to uptake micronutrients. Nitrogen is an essential source for biosynthetic pathways. Adaptation to nutritional stress is a key feature of fungi in host tissues. Fungi utilize nitrogen sources through Nitrogen Catabolite Repression (NCR). NCR ensures the scavenging, uptake and catabolism of alternative nitrogen sources, when preferential ones, such as glutamine or ammonium, are unavailable. The NanoUPLC-MSE proteomic approach was used to investigate the NCR response of Paracoccidioides lutzii after growth on proline or glutamine as a nitrogen source. A total of 338 differentially expressed proteins were identified. P. lutzii demonstrated that gluconeogenesis, β-oxidation, glyoxylate cycle, adhesin-like proteins, stress response and cell wall remodeling were triggered in NCR-proline conditions. In addition, within macrophages, yeast cells trained under NCR-proline conditions showed an increased ability to survive. In general, this study allows a comprehensive understanding of the NCR response employed by the fungus to overcome nutritional starvation, which in the human host is represented by nutritional immunity. In turn, the pathogen requires rapid adaptation to the changing microenvironment induced by macrophages to achieve successful infection.
Keywords: NCR-proline; Paracoccidioides; metabolic reprogramming; nitrogen starvation; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Shikanai-Yasuda M.A., Mendes R.P., Colombo A.L., Queiroz-Telles F.d., Kono A.S.G., Paniago A.M., Nathan A., Valle A.C.F.d., Bagagli E., Benard G. Brazilian guidelines for the clinical management of paracoccidioidomycosis. Rev. Soc. Bras. Med. Trop. 2017;50:715–740. doi: 10.1590/0037-8682-0230-2017. - DOI - PubMed
Grants and funding
- 478990/2012-0 and 408042/2021-4/National Council for Scientific and Technological Development
- INCT-IPH-FAPEG/Fundação de Apoio a Pesquisa do Estado de Goiás
- 308237/2022-6/National Council for Scientific and Technological Development
- DS fellowships/Coordenação de Aperfeicoamento de Pessoal de Nível Superior
LinkOut - more resources
Full Text Sources

