Deacetylation of ATG7 drives the induction of macroautophagy and LC3-associated microautophagy
- PMID: 37999993
- PMCID: PMC11135844
- DOI: 10.1080/15548627.2023.2287932
Deacetylation of ATG7 drives the induction of macroautophagy and LC3-associated microautophagy
Abstract
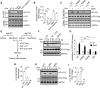
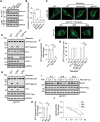
LC3 lipidation plays an important role in the regulation of macroautophagy and LC3-associated microautophagy. The E1-like enzyme ATG7 is one of the core components that are directly involved in LC3 lipidation reaction. Here, we provide evidence showing that acetylation of ATG7 tightly controls its enzyme activity to regulate the induction of macroautophagy and LC3-associated microautophagy. Mechanistically, acetylation of ATG7 disrupts its interaction with the E2-like enzyme ATG3, leading to an inhibition of LC3 lipidation in vitro and in vivo. Functionally, in response to various different stimuli, cellular ATG7 undergoes deacetylation to induce macroautophagy and LC3-associated microautophagy, which are necessary for cells to eliminate cytoplasmic DNA and degrade lysosome membrane proteins, respectively. Taken together, these findings reveal that ATG7 acetylation acts as a critical rheostat in controlling LC3 lipidation and related cellular processes.Abbreviations: AMPK: AMP-activated protein kinase; ATG: autophagy-related; cGAMP: cyclic GMP-AMP; CGAS: cyclic GMP-AMP synthase; CREBBP/CBP: CREB binding protein; EGF: epidermal growth factor; EGFR: epidermal growth factor receptor; EP300/p300: E1A binding protein p300; IFNB1: interferon beta 1; ISD: interferon stimulatory DNA; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MCOLN1/TRPML1: mucolipin TRP cation channel 1; MEF: mouse embryonic fibroblast; MTOR: mechanistic target of rapamycin kinase; NAM: nicotinamide; PE: phosphatidylethanolamine; PTM: post-translational modification; RB1CC1/FIP200: RB1 inducible coiled-coil 1; SIRT: sirtuin; SQSTM1/p62: sequestosome 1; STING1: stimulator of interferon response cGAMP interactor 1; TSA: trichostatin A; ULK1: unc-51 like autophagy activating kinase 1; WIPI2: WD repeat domain, phosphoinositide interacting 2; WT: wild-type.
Keywords: ATG7; Acetylation; LC3 lipidation; macroautophagy; microautophagy.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



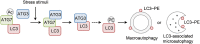
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
