Partial in vivo reprogramming enables injury-free intestinal regeneration via autonomous Ptgs1 induction
- PMID: 38000027
- PMCID: PMC10672161
- DOI: 10.1126/sciadv.adi8454
Partial in vivo reprogramming enables injury-free intestinal regeneration via autonomous Ptgs1 induction
Abstract
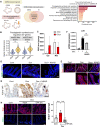
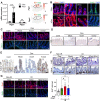
Tissue regeneration after injury involves the dedifferentiation of somatic cells, a natural adaptive reprogramming that leads to the emergence of injury-responsive cells with fetal-like characteristics. However, there is no direct evidence that adaptive reprogramming involves a shared molecular mechanism with direct cellular reprogramming. Here, we induced dedifferentiation of intestinal epithelial cells using OSKM (Oct4, Sox2, Klf4, and c-Myc) in vivo. The OSKM-induced forced dedifferentiation showed similar molecular features of intestinal regeneration, including a transition from homeostatic cell types to injury-responsive-like cell types. These injury-responsive-like cells, sharing gene signatures of revival stem cells and atrophy-induced villus epithelial cells, actively assisted tissue regeneration following damage. In contrast to normal intestinal regeneration involving Ptgs2 induction, the OSKM promotes autonomous production of prostaglandin E2 via epithelial Ptgs1 expression. These results indicate prostaglandin synthesis is a common mechanism for intestinal regeneration but involves a different enzyme when partial reprogramming is applied to the intestinal epithelium.
Figures




References
-
- S. H. Tan, P. Phuah, L. T. Tan, S. Yada, J. Goh, L. B. Tomaz, M. Chua, E. Wong, B. Lee, N. Barker, A constant pool of Lgr5(+) intestinal stem cells is required for intestinal homeostasis. Cell Rep. 34, 108633 (2021). - PubMed
-
- N. Barker, Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 15, 19–33 (2014). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

