Intestinal Paneth cell differentiation relies on asymmetric regulation of Wnt signaling by Daam1/2
- PMID: 38000028
- PMCID: PMC10672176
- DOI: 10.1126/sciadv.adh9673
Intestinal Paneth cell differentiation relies on asymmetric regulation of Wnt signaling by Daam1/2
Abstract
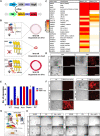
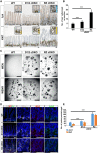
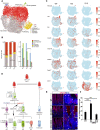
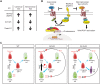
The mammalian intestine is one of the most rapidly self-renewing tissues, driven by stem cells residing at the crypt bottom. Paneth cells form a major element of the niche microenvironment providing various growth factors to orchestrate intestinal stem cell homeostasis, such as Wnt3. Different Wnt ligands can selectively activate β-catenin-dependent (canonical) or -independent (noncanonical) signaling. Here, we report that the Dishevelled-associated activator of morphogenesis 1 (Daam1) and its paralogue Daam2 asymmetrically regulate canonical and noncanonical Wnt (Wnt/PCP) signaling. Daam1/2 interacts with the Wnt inhibitor RNF43, and Daam1/2 double knockout stimulates canonical Wnt signaling by preventing RNF43-dependent degradation of the Wnt receptor, Frizzled (Fzd). Single-cell RNA sequencing analysis revealed that Paneth cell differentiation is impaired by Daam1/2 depletion because of defective Wnt/PCP signaling. Together, we identified Daam1/2 as an unexpected hub molecule coordinating both canonical and noncanonical Wnt, which is fundamental for specifying an adequate number of Paneth cells.
Figures




References
-
- Beumer J., Clevers H., Cell fate specification and differentiation in the adult mammalian intestine. Nat. Rev. Mol. Cell Biol. 22, 39–53 (2021). - PubMed
-
- Nusse R., Clevers H., Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017). - PubMed
-
- Koo B. K., Spit M., Jordens I., Low T. Y., Stange D. E., van de Wetering M., van Es J. H., Mohammed S., Heck A. J., Maurice M. M., Clevers H., Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488, 665–669 (2012). - PubMed
-
- Hao H. X., Xie Y., Zhang Y., Charlat O., Oster E., Avello M., Lei H., Mickanin C., Liu D., Ruffner H., Mao X., Ma Q., Zamponi R., Bouwmeester T., Finan P. M., Kirschner M. W., Porter J. A., Serluca F. C., Cong F., ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485, 195–200 (2012). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

