SARS-CoV-2 breakthrough infections following inactivated vaccine vaccination induce few neutralizing antibodies against the currently emerging Omicron XBB variants
- PMID: 38000528
- PMCID: PMC10877414
- DOI: 10.1016/j.virs.2023.11.007
SARS-CoV-2 breakthrough infections following inactivated vaccine vaccination induce few neutralizing antibodies against the currently emerging Omicron XBB variants
Abstract
- •
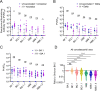
Inactivated vaccine breakthrough infection with ancestral variants induced nearly undetectable nAbs against XBB variants.
- •
Inactivated vaccine breakthrough infection with Omicron BA.1 or BA.5 evoked very weak nAbs against XBB variants.
- •
BA.5 infection induced higher nAbs against XBB variants than BA.1 infection.
Figures

Similar articles
-
Repeated vaccination of inactivated SARS-CoV-2 vaccine dampens neutralizing antibodies against Omicron variants in breakthrough infection.Cell Res. 2023 Mar;33(3):258-261. doi: 10.1038/s41422-023-00781-8. Epub 2023 Jan 25. Cell Res. 2023. PMID: 36697703 Free PMC article. No abstract available.
-
A mosaic-type trimeric RBD-based COVID-19 vaccine candidate induces potent neutralization against Omicron and other SARS-CoV-2 variants.Elife. 2022 Aug 25;11:e78633. doi: 10.7554/eLife.78633. Elife. 2022. PMID: 36004719 Free PMC article.
-
Evaluation of Neutralizing Activity against Omicron Subvariants in BA.5 Breakthrough Infection and Three-Dose Vaccination Using a Novel Chemiluminescence-Based, Virus-Mediated Cytopathic Assay.Microbiol Spectr. 2023 Aug 17;11(4):e0066023. doi: 10.1128/spectrum.00660-23. Epub 2023 Jun 13. Microbiol Spectr. 2023. PMID: 37310218 Free PMC article.
-
Previous exposure to Spike-providing parental strains confers neutralizing immunity to XBB lineage and other SARS-CoV-2 recombinants in the context of vaccination.Emerg Microbes Infect. 2023 Dec;12(2):2270071. doi: 10.1080/22221751.2023.2270071. Epub 2023 Oct 31. Emerg Microbes Infect. 2023. PMID: 37869789 Free PMC article.
-
Variants of SARS-CoV-2: Influences on the Vaccines' Effectiveness and Possible Strategies to Overcome Their Consequences.Medicina (Kaunas). 2023 Mar 5;59(3):507. doi: 10.3390/medicina59030507. Medicina (Kaunas). 2023. PMID: 36984508 Free PMC article. Review.
Cited by
-
Single-cell analysis unravels the role of NK cells and monocytes in the control of SARS-CoV-2 breakthrough infections in vaccinated individuals.Hum Vaccin Immunother. 2025 Dec;21(1):2534219. doi: 10.1080/21645515.2025.2534219. Epub 2025 Jul 29. Hum Vaccin Immunother. 2025. PMID: 40729342 Free PMC article.
References
-
- Arunachalam P.S., Lai L., Samaha H., Feng Y., Hu M., Hui H.S., Wali B., Ellis M., Davis-Gardner M.E., Huerta C., Bechnak K., Bechnak S., Lee M., Litvack M.B., Losada C., Grifoni A., Sette A., Zarnitsyna V.I., Rouphael N., Suthar M.S., Pulendran B. Durability of immune responses to mRNA booster vaccination against COVID-19. J. Clin. Invest. 2023;133 - PMC - PubMed
-
- Devasundaram S., Terpos E., Rosati M., Ntanasis-Stathopoulos I., Bear J., Burns R., Skourti S., Malandrakis P., Trougakos I.P., Dimopoulos M.A., Pavlakis G.N., Felber B.K. XBB.1.5 neutralizing antibodies upon bivalent COVID-19 vaccination are similar to XBB but lower than BQ.1.1. Am. J. Hematol. 2023;98:E123–E126. - PMC - PubMed
-
- Hoffmann M., Arora P., Nehlmeier I., Kempf A., Cossmann A., Schulz S.R., Morillas Ramos G., Manthey L.A., Jäck H.M., Behrens G.M.N., Pöhlmann S. Profound neutralization evasion and augmented host cell entry are hallmarks of the fast-spreading SARS-CoV-2 lineage XBB.1.5. Cell. Mol. Immunol. 2023;20:419–422. - PMC - PubMed
-
- Kaku Y., Kosugi Y., Uriu K., Ito J., Hinay A.A., Jr., Kuramochi J., Sadamasu K., Yoshimura K., Asakura H., Nagashima M., Genotype to Phenotype Japan (G2P-Japan) Consortium, Sato, K Antiviral efficacy of the SARS-CoV-2 XBB breakthrough infection sera against omicron subvariants including EG.5. Lancet Infect. Dis. 2023;23:e395–e396. - PubMed
-
- Ma K.C., Shirk P., Lambrou A.S., Hassell N., Zheng X.Y., Payne A.B., Ali A.R., Batra D., Caravas J., Chau R., Cook P.W., Howard D., Kovacs N.A., Lacek K.A., Lee J.S., MacCannell D.R., Malapati L., Mathew S., Mittal N., Nagilla R.R., Parikh R., Paul P., Rambo-Martin B.L., Shepard S.S., Sheth M., Wentworth D.E., Winn A., Hall A.J., Silk B.J., Thornburg N., Kondor R., Scobie H.M., Paden C.R. Genomic surveillance for SARS-CoV-2 variants: circulation of omicron lineages - United States, january 2022-may 2023. MMWR Morb. Mortal. Wkly. Rep. 2023;72:651–656. - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

