Genetic separation of Brca1 functions reveal mutation-dependent Polθ vulnerabilities
- PMID: 38001070
- PMCID: PMC10673838
- DOI: 10.1038/s41467-023-43446-1
Genetic separation of Brca1 functions reveal mutation-dependent Polθ vulnerabilities
Abstract
Homologous recombination (HR)-deficiency induces a dependency on DNA polymerase theta (Polθ/Polq)-mediated end joining, and Polθ inhibitors (Polθi) are in development for cancer therapy. BRCA1 and BRCA2 deficient cells are thought to be synthetic lethal with Polθ, but whether distinct HR gene mutations give rise to equivalent Polθ-dependence, and the events that drive lethality, are unclear. In this study, we utilized mouse models with separate Brca1 functional defects to mechanistically define Brca1-Polθ synthetic lethality. Surprisingly, homozygous Brca1 mutant, Polq-/- cells were viable, but grew slowly and had chromosomal instability. Brca1 mutant cells proficient in DNA end resection were significantly more dependent on Polθ for viability; here, treatment with Polθi elevated RPA foci, which persisted through mitosis. In an isogenic system, BRCA1 null cells were defective, but PALB2 and BRCA2 mutant cells exhibited active resection, and consequently stronger sensitivity to Polθi. Thus, DNA end resection is a critical determinant of Polθi sensitivity in HR-deficient cells, and should be considered when selecting patients for clinical studies.
© 2023. The Author(s).
Conflict of interest statement
G.P.G. has received research funding from Breakpoint Therapeutics and Merck. R.T.P. is a cofounder and chief scientific officer of Recombination Therapeutics, LLC. The remaining authors declare no competing interests.
Figures


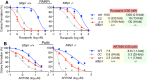
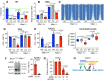


References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

