Identification of peptides from honeybee gut symbionts as potential antimicrobial agents against Melissococcus plutonius
- PMID: 38001079
- PMCID: PMC10673953
- DOI: 10.1038/s41467-023-43352-6
Identification of peptides from honeybee gut symbionts as potential antimicrobial agents against Melissococcus plutonius
Abstract
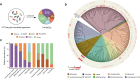
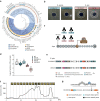
Eusocial pollinators are crucial elements in global agriculture. The honeybees and bumblebees are associated with a simple yet host-restricted gut community, which protect the hosts against pathogen infections. Recent genome mining has led to the discovery of biosynthesis pathways of bioactive natural products mediating microbe-microbe interactions from the gut microbiota. Here, we investigate the diversity of biosynthetic gene clusters in the bee gut microbiota by analyzing 477 genomes from cultivated bacteria and metagenome-assembled genomes. We identify 744 biosynthetic gene clusters (BGCs) covering multiple chemical classes. While gene clusters for the post-translationally modified peptides are widely distributed in the bee guts, the distribution of the BGC classes varies significantly in different bee species among geographic locations, which is attributed to the strain-level variation of bee gut members in the chemical repertoire. Interestingly, we find that Gilliamella strains possessing a thiopeptide-like BGC show potent activity against the pathogenic Melissococcus plutonius. The spectrometry-guided genome mining reveals a RiPP-encoding BGC from Gilliamella with a 10 amino acid-long core peptide exhibiting antibacterial potentials. This study illustrates the widespread small-molecule-encoding BGCs in the bee gut symbionts and provides insights into the bacteria-derived natural products as potential antimicrobial agents against pathogenic infections.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Mining metagenomic data to gain a new insight into the gut microbial biosynthetic potential in placental mammals.Microbiol Spectr. 2024 Oct 3;12(10):e0086424. doi: 10.1128/spectrum.00864-24. Epub 2024 Aug 20. Microbiol Spectr. 2024. PMID: 39162518 Free PMC article.
-
Specific Strains of Honeybee Gut Lactobacillus Stimulate Host Immune System to Protect against Pathogenic Hafnia alvei.Microbiol Spectr. 2022 Feb 23;10(1):e0189621. doi: 10.1128/spectrum.01896-21. Epub 2022 Jan 5. Microbiol Spectr. 2022. PMID: 34985299 Free PMC article.
-
Distinct Roles of Honeybee Gut Bacteria on Host Metabolism and Neurological Processes.Microbiol Spectr. 2022 Apr 27;10(2):e0243821. doi: 10.1128/spectrum.02438-21. Epub 2022 Mar 10. Microbiol Spectr. 2022. PMID: 35266810 Free PMC article.
-
Functional and evolutionary insights into the simple yet specific gut microbiota of the honey bee from metagenomic analysis.Gut Microbes. 2013 Jan-Feb;4(1):60-5. doi: 10.4161/gmic.22517. Epub 2012 Oct 12. Gut Microbes. 2013. PMID: 23060052 Free PMC article. Review.
-
Comparative Metagenomic Analysis of Biosynthetic Diversity across Sponge Microbiomes Highlights Metabolic Novelty, Conservation, and Diversification.mSystems. 2022 Aug 30;7(4):e0035722. doi: 10.1128/msystems.00357-22. Epub 2022 Jul 18. mSystems. 2022. PMID: 35862823 Free PMC article. Review.
Cited by
-
Phylogenetic and functional diversity among Drosophila-associated metagenome-assembled genomes.mSystems. 2025 Jul 22;10(7):e0002725. doi: 10.1128/msystems.00027-25. Epub 2025 Jul 2. mSystems. 2025. PMID: 40600712 Free PMC article.
-
Screening and identification of antimicrobial peptides from the gut microbiome of cockroach Blattella germanica.Microbiome. 2024 Dec 21;12(1):272. doi: 10.1186/s40168-024-01985-9. Microbiome. 2024. PMID: 39709489 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

