Tamoxifen for the treatment of myeloproliferative neoplasms: A Phase II clinical trial and exploratory analysis
- PMID: 38001082
- PMCID: PMC10673935
- DOI: 10.1038/s41467-023-43175-5
Tamoxifen for the treatment of myeloproliferative neoplasms: A Phase II clinical trial and exploratory analysis
Abstract
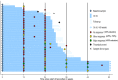
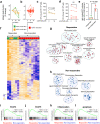
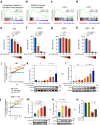
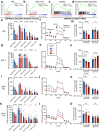
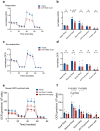
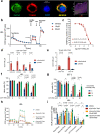
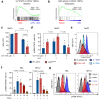
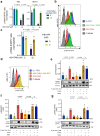
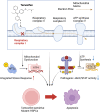
Current therapies for myeloproliferative neoplasms (MPNs) improve symptoms but have limited effect on tumor size. In preclinical studies, tamoxifen restored normal apoptosis in mutated hematopoietic stem/progenitor cells (HSPCs). TAMARIN Phase-II, multicenter, single-arm clinical trial assessed tamoxifen's safety and activity in patients with stable MPNs, no prior thrombotic events and mutated JAK2V617F, CALRins5 or CALRdel52 peripheral blood allele burden ≥20% (EudraCT 2015-005497-38). 38 patients were recruited over 112w and 32 completed 24w-treatment. The study's A'herns success criteria were met as the primary outcome ( ≥ 50% reduction in mutant allele burden at 24w) was observed in 3/38 patients. Secondary outcomes included ≥25% reduction at 24w (5/38), ≥50% reduction at 12w (0/38), thrombotic events (2/38), toxicities, hematological response, proportion of patients in each IWG-MRT response category and ELN response criteria. As exploratory outcomes, baseline analysis of HSPC transcriptome segregates responders and non-responders, suggesting a predictive signature. In responder HSPCs, longitudinal analysis shows high baseline expression of JAK-STAT signaling and oxidative phosphorylation genes, which are downregulated by tamoxifen. We further demonstrate in preclinical studies that in JAK2V617F+ cells, 4-hydroxytamoxifen inhibits mitochondrial complex-I, activates integrated stress response and decreases pathogenic JAK2-signaling. These results warrant further investigation of tamoxifen in MPN, with careful consideration of thrombotic risk.
© 2023. The Author(s).
Conflict of interest statement
C.N.H. reports funded research from Novartis; speaker fees from Novartis, Janssen, CTI, Celgene, and Medscape; and advisory board membership for Incyte, CTI, Sierra Oncology, Novartis, Celgene, Roche, AOP pharma, Geron, and AstraZeneca. A.J.M. reports funded research from Novartis, Celgene/BMS and consultancy for Abbvie, CTI, and Gilead. S.K. reports funded research and advisory board membership for Novartis. J.E. reports advisory board membership for Novartis, Incyte and Celgene/BMS. M.F.M. reports consultancies for Celgene and BMS; and advisory board membership for Novartis, Jazz, and Abbvie. S.N. reports speaker fees for Takeda, Celgene, Novartis, MSD and Alexion. D.M. reports consultancy for Incyte, Pfizer, Novartis, and Bristol-Myers Squibb. M.W.D. reports funded research from Blueprint Medicine Corporation and advisory board membership for Bristol-Myers Squibb, Novartis, Gilead, Pfizer, Jazz, Takeda, and Astellas. The remaining authors declare no competing financial interests. A.L.G. reports speaker fees from Novartis and advisory board membership AOP pharma and BMS.
Figures
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

