Stepwise differentiation of follicular helper T cells reveals distinct developmental and functional states
- PMID: 38001088
- PMCID: PMC10674016
- DOI: 10.1038/s41467-023-43427-4
Stepwise differentiation of follicular helper T cells reveals distinct developmental and functional states
Abstract
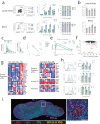
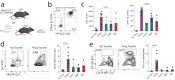
Follicular helper T (Tfh) cells are essential for the formation of high affinity antibodies after vaccination or infection. Although the signals responsible for initiating Tfh differentiation from naïve T cells have been studied, the signals controlling sequential developmental stages culminating in optimal effector function are not well understood. Here we use fate mapping strategies for the cytokine IL-21 to uncover sequential developmental stages of Tfh differentiation including a progenitor-like stage, a fully developed effector stage and a post-effector Tfh stage that maintains transcriptional and epigenetic features without IL-21 production. We find that progression through these stages are controlled intrinsically by the transcription factor FoxP1 and extrinsically by follicular regulatory T cells. Through selective deletion of Tfh stages, we show that these cells control antibody dynamics during distinct stages of the germinal center reaction in response to a SARS-CoV-2 vaccine. Together, these studies demonstrate the sequential phases of Tfh development and how they promote humoral immunity.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Wing, J. B., Lim, E. L. & Sakaguchi, S. Control of foreign Ag-specific Ab responses by Treg and Tfr. Immunol. Rev.296, 104–119 (2020). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

