Conjugation of HIV-1 envelope to hepatitis B surface antigen alters vaccine responses in rhesus macaques
- PMID: 38001122
- PMCID: PMC10673864
- DOI: 10.1038/s41541-023-00775-y
Conjugation of HIV-1 envelope to hepatitis B surface antigen alters vaccine responses in rhesus macaques
Abstract
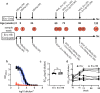
An effective HIV-1 vaccine remains a critical unmet need for ending the AIDS epidemic. Vaccine trials conducted to date have suggested the need to increase the durability and functionality of vaccine-elicited antibodies to improve efficacy. We hypothesized that a conjugate vaccine based on the learned response to immunization with hepatitis B virus could be utilized to expand T cell help and improve antibody production against HIV-1. To test this, we developed an innovative conjugate vaccine regimen that used a modified vaccinia virus Ankara (MVA) co-expressing HIV-1 envelope (Env) and the hepatitis B virus surface antigen (HBsAg) as a prime, followed by two Env-HBsAg conjugate protein boosts. We compared the immunogenicity of this conjugate regimen to matched HIV-1 Env-only vaccines in two groups of 5 juvenile rhesus macaques previously immunized with hepatitis B vaccines in infancy. We found expansion of both HIV-1 and HBsAg-specific circulating T follicular helper cells and elevated serum levels of CXCL13, a marker for germinal center activity, after boosting with HBsAg-Env conjugate antigens in comparison to Env alone. The conjugate vaccine elicited higher levels of antibodies binding to select HIV Env antigens, but we did not observe significant improvement in antibody functionality, durability, maturation, or B cell clonal expansion. These data suggests that conjugate vaccination can engage both HIV-1 Env and HBsAg specific T cell help and modify antibody responses at early time points, but more research is needed to understand how to leverage this strategy to improve the durability and efficacy of next-generation HIV vaccines.
© 2023. The Author(s).
Conflict of interest statement
S.R.P. provides individual consulting services to Moderna, Merck, Dynavax, Pfizer, GlaxoSmithKline (CMV vaccines) and HOOKIPA Biotech GmbH. Merck Vaccines and Moderna have provided grants for her Institutional sponsored programs.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources

