LUBAC is required for RIG-I sensing of RNA viruses
- PMID: 38001254
- PMCID: PMC10781740
- DOI: 10.1038/s41418-023-01233-x
LUBAC is required for RIG-I sensing of RNA viruses
Abstract
The ability of cells to mount an interferon response to virus infections depends on intracellular nucleic acid sensing pattern recognition receptors (PRRs). RIG-I is an intracellular PRR that binds short double-stranded viral RNAs to trigger MAVS-dependent signalling. The RIG-I/MAVS signalling complex requires the coordinated activity of multiple kinases and E3 ubiquitin ligases to activate the transcription factors that drive type I and type III interferon production from infected cells. The linear ubiquitin chain assembly complex (LUBAC) regulates the activity of multiple receptor signalling pathways in both ligase-dependent and -independent ways. Here, we show that the three proteins that constitute LUBAC have separate functions in regulating RIG-I signalling. Both HOIP, the E3 ligase capable of generating M1-ubiquitin chains, and LUBAC accessory protein HOIL-1 are required for viral RNA sensing by RIG-I. The third LUBAC component, SHARPIN, is not required for RIG-I signalling. These data cement the role of LUBAC as a positive regulator of RIG-I signalling and as an important component of antiviral innate immune responses.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
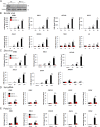

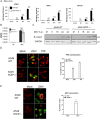

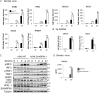

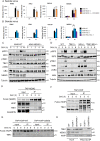
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

