Transcriptomic profiling of cerebrospinal fluid identifies ALS pathway enrichment and RNA biomarkers in MND individuals
- PMID: 38001563
- PMCID: PMC10903246
- DOI: 10.1177/15353702231209427
Transcriptomic profiling of cerebrospinal fluid identifies ALS pathway enrichment and RNA biomarkers in MND individuals
Abstract
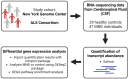
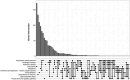
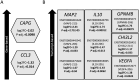
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disorder and the most common form of motor neurone disease (MND) which is characterized by the damage and death of motor neurons in the brain and spinal cord of affected individuals. Due to the heterogeneity of the disease, a better understanding of the interaction between genetics and biochemistry with the identification of biomarkers is crucial for therapy development. In this study, we used cerebrospinal fluid (CSF) RNA-sequencing data from the New York Genome Center (NYGC) ALS Consortium and analyzed differential gene expression between 47 MND individuals and 29 healthy controls. Pathway analysis showed that the affected genes are enriched in many pathways associated with ALS, including nucleocytoplasmic transport, autophagy, and apoptosis. Moreover, we assessed differential expression on both gene- and transcript-based levels and demonstrate that the expression of previously identified potential biomarkers, including CAPG, CCL3, and MAP2, was significantly higher in MND individuals. Ultimately, this study highlights the transcriptomic composition of CSF which enables insights into changes in the brain in ALS and therefore increases the confidence in the use of CSF for biomarker development.
Keywords: Cerebrospinal fluid; RNA-seq; amyotrophic lateral sclerosis; biomarker; motor neurone disease; transcriptome.
Conflict of interest statement
Declaration of conflicting interestsThe author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures
Similar articles
-
Proteomics in cerebrospinal fluid and spinal cord suggests UCHL1, MAP2 and GPNMB as biomarkers and underpins importance of transcriptional pathways in amyotrophic lateral sclerosis.Acta Neuropathol. 2020 Jan;139(1):119-134. doi: 10.1007/s00401-019-02093-x. Epub 2019 Nov 7. Acta Neuropathol. 2020. PMID: 31701227
-
Profiling non-coding RNA expression in cerebrospinal fluid of amyotrophic lateral sclerosis patients.Ann Med. 2022 Dec;54(1):3069-3078. doi: 10.1080/07853890.2022.2138530. Ann Med. 2022. PMID: 36314539 Free PMC article.
-
Diagnostic and prognostic value of CSF neurofilaments in a cohort of patients with motor neuron disease: A cross-sectional study.J Cell Mol Med. 2021 Apr;25(8):3765-3771. doi: 10.1111/jcmm.16240. Epub 2021 Feb 20. J Cell Mol Med. 2021. PMID: 33609080 Free PMC article.
-
New insights into the gene expression associated to amyotrophic lateral sclerosis.Life Sci. 2018 Jan 15;193:110-123. doi: 10.1016/j.lfs.2017.12.016. Epub 2017 Dec 11. Life Sci. 2018. PMID: 29241710 Review.
-
Profiling TREM2 expression in amyotrophic lateral sclerosis.Brain Behav Immun. 2023 Mar;109:117-126. doi: 10.1016/j.bbi.2023.01.013. Epub 2023 Jan 18. Brain Behav Immun. 2023. PMID: 36681358 Review.
Cited by
-
Molecular adaptations underlying high-frequency hearing in the brain of CF bats species.BMC Genomics. 2024 Mar 16;25(1):279. doi: 10.1186/s12864-024-10212-6. BMC Genomics. 2024. PMID: 38493092 Free PMC article.
-
Pathophysiology, Clinical Heterogeneity, and Therapeutic Advances in Amyotrophic Lateral Sclerosis: A Comprehensive Review of Molecular Mechanisms, Diagnostic Challenges, and Multidisciplinary Management Strategies.Life (Basel). 2025 Apr 14;15(4):647. doi: 10.3390/life15040647. Life (Basel). 2025. PMID: 40283201 Free PMC article. Review.
-
Whole blood transcriptome profile identifies motor neurone disease RNA biomarker signatures.Exp Biol Med (Maywood). 2025 Jan 8;249:10401. doi: 10.3389/ebm.2024.10401. eCollection 2024. Exp Biol Med (Maywood). 2025. PMID: 39844875 Free PMC article.
-
Phenotypical Characterization of C9ALS Patients from the Emilia Romagna Registry of ALS: A Retrospective Case-Control Study.Genes (Basel). 2025 Mar 4;16(3):309. doi: 10.3390/genes16030309. Genes (Basel). 2025. PMID: 40149460 Free PMC article.
-
Immunological Fluid Biomarkers in Frontotemporal Dementia: A Systematic Review.Biomolecules. 2025 Mar 24;15(4):473. doi: 10.3390/biom15040473. Biomolecules. 2025. PMID: 40305176 Free PMC article.
References
-
- Ajroud-Driss S, Siddique T. Sporadic and hereditary amyotrophic lateral sclerosis (ALS). Biochim Biophys Acta 2015;1852:679–84 - PubMed
-
- Shatunov A, Al-Chalabi A. The genetic architecture of ALS. Neurobiol Dis 2021;147:105156. - PubMed
-
- Veldink JH. ALS genetic epidemiology “how simplex is the genetic epidemiology of ALS?” J Neurol Neurosurg Psychiatry 2017;88:537. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

