Prognostic Potential of Galectin-9 mRNA Expression in Chronic Lymphocytic Leukemia
- PMID: 38001630
- PMCID: PMC10670166
- DOI: 10.3390/cancers15225370
Prognostic Potential of Galectin-9 mRNA Expression in Chronic Lymphocytic Leukemia
Abstract
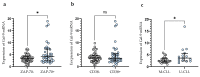
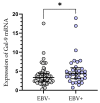
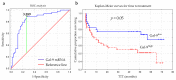
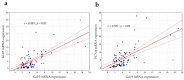
Galectin-9 (Gal-9), very poorly characterized in chronic lymphocytic leukemia (CLL), was chosen in our study to examine its potential role as a CLL biomarker. The relation of Gal-9 expression in malignant B-cells and other routinely measured CLL markers, as well as its clinical relevance are poorly understood. Gal-9 mRNA expression was quantified with RT-qPCR in purified CD19+ B-cells of 100 CLL patients and analyzed in the context of existing clinical data. Our results revealed the upregulation of Gal-9 mRNA in CLL cells. High Gal-9 mRNA expression was closely associated with unfavorable prognostic markers. In addition, Gal-9 expression in leukemic cells was significantly elevated in CLL patients who did not respond to the first-line therapy compared to those who did respond. This suggests its potential predictive value. Importantly, Gal-9 was an independent predictor for the time to treatment parameters. Thus, we can suggest an adverse role of Gal-9 expression in CLL. Interestingly, it is possible that Gal-9 expression is induced in B-cells by EBV infection, so we determined the patients' EBV status. Our suggestion is that EBV coinfection could worsen prognosis in CLL, partly due to Gal-9 expression upregulation caused by EBV.
Keywords: CLL; EBV; Galectin-9; biomarker; prognostic markers.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

