An Exploration of the Inhibitory Mechanism of Rationally Screened Benzofuran-1,3,4-Oxadiazoles and-1,2,4-Triazoles as Inhibitors of NS5B RdRp Hepatitis C Virus through Pharmacoinformatic Approaches
- PMID: 38002085
- PMCID: PMC10669698
- DOI: 10.3390/biomedicines11113085
An Exploration of the Inhibitory Mechanism of Rationally Screened Benzofuran-1,3,4-Oxadiazoles and-1,2,4-Triazoles as Inhibitors of NS5B RdRp Hepatitis C Virus through Pharmacoinformatic Approaches
Abstract
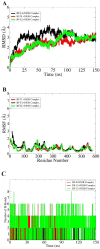
Benzofuran, 1,3,4-oxadiazole, and 1,2,4-triazole are privileged heterocyclic moieties that display the most promising and wide spectrum of biological activities against a wide variety of diseases. In the current study, benzofuran-1,3,4-oxadiazole BF1-BF7 and benzofuran-1,2,4-triazole compounds BF8-BF15 were tested against HCV NS5B RNA-dependent RNA polymerase (RdRp) utilizing structure-based screening via a computer-aided drug design (CADD) approach. A molecular docking approach was applied to evaluate the binding potential of benzofuran-appended 1,3,4-oxadiazole and 1,2,4-triazole BF1-BF15 molecules. Benzofuran-1,3,4-oxadiazole scaffolds BF1-BF7 showed lesser binding affinities (-12.63 to -14.04 Kcal/mol) than benzofuran-1,2,4-triazole scaffolds BF8-BF15 (-14.11 to -16.09 Kcal/mol) against the HCV NS5B enzyme. Molecular docking studies revealed the excellent binding affinity scores exhibited by benzofuran-1,2,4-triazole structural motifs BF-9 (-16.09 Kcal/mol), BF-12 (-15.75 Kcal/mol), and BF-13 (-15.82 Kcal/mol), respectively, which were comparatively better than benzofuran-based HCV NS5B inhibitors' standard reference drug Nesbuvir (-15.42 Kcal/mol). A molecular dynamics simulation assay was also conducted to obtain valuable insights about the enzyme-compounds interaction profile and structural stability, which indicated the strong intermolecular energies of the BF-9+NS5B complex and the BF-12+NS5B complex as per the MM-PBSA method, while the BF-12+NS5B complex was the most stable system as per the MM-GBSA calculation. The drug-likeness and ADMET studies of all the benzofuran-1,2,4-triazole derivatives BF8-BF15 revealed that these compounds possessed good medicinal chemistry profiles in agreement with all the evaluated parameters for being drugs. The molecular docking affinity scores, MM-PBSA/MM-GBSA and MD-simulation stability analysis, drug-likeness profiling, and ADMET study assessment indicated that N-4-fluorophenyl-S-linked benzofuran-1,2,4-triazole BF-12 could be a future promising anti-HCV NS5B RdRp inhibitor therapeutic drug candidate that has a structural agreement with the Nesbuvir standard reference drug.
Keywords: ADMET studies; DFT studies; MD simulations; MM-PBSA; RdRp NS5B inhibitors; SAR; benzofuran derivatives; energy decomposition analysis; hepatitis C; molecular docking.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









Similar articles
-
In Silico Development of Novel Benzofuran-1,3,4-Oxadiazoles as Lead Inhibitors of M. tuberculosis Polyketide Synthase 13.Pharmaceuticals (Basel). 2023 Jun 1;16(6):829. doi: 10.3390/ph16060829. Pharmaceuticals (Basel). 2023. PMID: 37375776 Free PMC article.
-
Design, Synthesis and Biological Exploration of Novel N-(9-Ethyl-9H-Carbazol-3-yl)Acetamide-Linked Benzofuran-1,2,4-Triazoles as Anti-SARS-CoV-2 Agents: Combined Wet/Dry Approach Targeting Main Protease (Mpro), Spike Glycoprotein and RdRp.Int J Mol Sci. 2024 Nov 26;25(23):12708. doi: 10.3390/ijms252312708. Int J Mol Sci. 2024. PMID: 39684420 Free PMC article.
-
Repositioning of RdRp Inhibitors Against HCV NS5B Polymerase Utilizing Structure-Based Molecular Docking.Comb Chem High Throughput Screen. 2022;25(4):702-719. doi: 10.2174/1386207324666210121111921. Comb Chem High Throughput Screen. 2022. PMID: 33475069
-
Small molecule NS5B RdRp non-nucleoside inhibitors for the treatment of HCV infection: A medicinal chemistry perspective.Eur J Med Chem. 2022 Oct 5;240:114595. doi: 10.1016/j.ejmech.2022.114595. Epub 2022 Jul 8. Eur J Med Chem. 2022. PMID: 35868125 Review.
-
Chemistry and Therapeutic Aspect of Triazole: Insight into the Structure-activity Relationship.Curr Pharm Des. 2023;29(34):2702-2720. doi: 10.2174/0113816128271288231023045049. Curr Pharm Des. 2023. PMID: 37916492 Review.
Cited by
-
A Comprehensive Review on Benzofuran Synthesis Featuring Innovative and Catalytic Strategies.ACS Omega. 2024 May 6;9(19):20728-20752. doi: 10.1021/acsomega.4c02677. eCollection 2024 May 14. ACS Omega. 2024. PMID: 38764672 Free PMC article. Review.
References
-
- Payne S. Viruses. Academic Press; Cambridge, MA, USA: 2017. Introduction to animal viruses; pp. 1–11. - DOI
-
- World Health Organization . Global Hepatitis Report. World Health Organization; Geneva, Switzerland: 2017.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

