Analysis of Structural Changes in the Protein near the Phosphorylation Site
- PMID: 38002246
- PMCID: PMC10668964
- DOI: 10.3390/biom13111564
Analysis of Structural Changes in the Protein near the Phosphorylation Site
Abstract
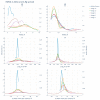
Modification of the protein after synthesis (PTM) often affects protein function as supported by numerous studies. However, there is no consensus about the degree of structural protein changes after modification. For phosphorylation of serine, threonine, and tyrosine, which is a common PTM in the biology of living organisms, we consider topical issues related to changes in the geometric parameters of a protein (Rg, RMSD, Cα displacement, SASA). The effect of phosphorylation on protein geometry was studied both for the whole protein and at the local level (i.e., in different neighborhoods of the modification site). Heterogeneity in the degree of protein structural changes after phosphorylation was revealed, which allowed for us to isolate a group of proteins having pronounced local structural changes in the neighborhoods of up to 15 amino acid residues from the modification site. This is a comparative study of protein structural changes in neighborhoods of 3-15 amino acid residues from the modified site. Amino acid phosphorylation in proteins with pronounced local changes caused switching from the inactive functional state to the active one.
Keywords: local change in protein structure; phosphorylation; post-translational modification; three-dimensional protein structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




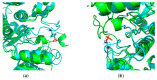
Similar articles
-
A novel network-based computational method to predict protein phosphorylation on tyrosine sites.J Bioinform Comput Biol. 2015 Dec;13(6):1542005. doi: 10.1142/S0219720015420056. J Bioinform Comput Biol. 2015. PMID: 26781824
-
Random coil chemical shifts for serine, threonine and tyrosine phosphorylation over a broad pH range.J Biomol NMR. 2019 Dec;73(12):713-725. doi: 10.1007/s10858-019-00283-z. Epub 2019 Oct 9. J Biomol NMR. 2019. PMID: 31598803 Free PMC article.
-
Cell signaling, post-translational protein modifications and NMR spectroscopy.J Biomol NMR. 2012 Nov;54(3):217-36. doi: 10.1007/s10858-012-9674-x. Epub 2012 Sep 26. J Biomol NMR. 2012. PMID: 23011410 Free PMC article.
-
Chemical Approaches to Studying Labile Amino Acid Phosphorylation.Top Curr Chem (Cham). 2017 Apr;375(2):22. doi: 10.1007/s41061-017-0111-1. Epub 2017 Feb 6. Top Curr Chem (Cham). 2017. PMID: 28168647 Review.
-
Protein Modification and Autophagy Activation.Adv Exp Med Biol. 2019;1206:237-259. doi: 10.1007/978-981-15-0602-4_12. Adv Exp Med Biol. 2019. PMID: 31776989 Review.
Cited by
-
Normal Mode Analysis Elicits Conformational Shifts in Proteins at Both Proximal and Distal Regions to the Phosphosite Stemming from Single-Site Phosphorylation.ACS Omega. 2024 May 30;9(23):24520-24537. doi: 10.1021/acsomega.4c00523. eCollection 2024 Jun 11. ACS Omega. 2024. PMID: 38882086 Free PMC article.
-
Plasma proteome fingerprint in kidney diseases.Front Mol Biosci. 2025 Jan 17;11:1494779. doi: 10.3389/fmolb.2024.1494779. eCollection 2024. Front Mol Biosci. 2025. PMID: 39896931 Free PMC article.
-
Extended range proteomic analysis of blood plasma from schizophrenia patients.Front Mol Biosci. 2024 Nov 21;11:1483933. doi: 10.3389/fmolb.2024.1483933. eCollection 2024. Front Mol Biosci. 2024. PMID: 39640846 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

