Neuroproteomic Analysis after SARS-CoV-2 Infection Reveals Overrepresented Neurodegeneration Pathways and Disrupted Metabolic Pathways
- PMID: 38002279
- PMCID: PMC10669333
- DOI: 10.3390/biom13111597
Neuroproteomic Analysis after SARS-CoV-2 Infection Reveals Overrepresented Neurodegeneration Pathways and Disrupted Metabolic Pathways
Abstract
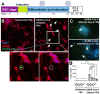
Besides respiratory illness, SARS-CoV-2, the causative agent of COVID-19, leads to neurological symptoms. The molecular mechanisms leading to neuropathology after SARS-CoV-2 infection are sparsely explored. SARS-CoV-2 enters human cells via different receptors, including ACE-2, TMPRSS2, and TMEM106B. In this study, we used a human-induced pluripotent stem cell-derived neuronal model, which expresses ACE-2, TMPRSS2, TMEM106B, and other possible SARS-CoV-2 receptors, to evaluate its susceptibility to SARS-CoV-2 infection. The neurons were exposed to SARS-CoV-2, followed by RT-qPCR, immunocytochemistry, and proteomic analyses of the infected neurons. Our findings showed that SARS-CoV-2 infects neurons at a lower rate than other human cells; however, the virus could not replicate or produce infectious virions in this neuronal model. Despite the aborted SARS-CoV-2 replication, the infected neuronal nuclei showed irregular morphology compared to other human cells. Since cytokine storm is a significant effect of SARS-CoV-2 infection in COVID-19 patients, in addition to the direct neuronal infection, the neurons were treated with pre-conditioned media from SARS-CoV-2-infected lung cells, and the neuroproteomic changes were investigated. The limited SARS-CoV-2 infection in the neurons and the neurons treated with the pre-conditioned media showed changes in the neuroproteomic profile, particularly affecting mitochondrial proteins and apoptotic and metabolic pathways, which may lead to the development of neurological complications. The findings from our study uncover a possible mechanism behind SARS-CoV-2-mediated neuropathology that might contribute to the lingering effects of the virus on the human brain.
Keywords: COVID-19; NeuroCOVID; SARS-CoV-2; apoptosis; iPSC-derived human neurons and astrocytes; mass spectrometry; metabolism; neurodegeneration; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
SARS-CoV-2 Infection of Human Neurons Is TMPRSS2 Independent, Requires Endosomal Cell Entry, and Can Be Blocked by Inhibitors of Host Phosphoinositol-5 Kinase.J Virol. 2023 Apr 27;97(4):e0014423. doi: 10.1128/jvi.00144-23. Epub 2023 Apr 11. J Virol. 2023. PMID: 37039676 Free PMC article.
-
Replication Kinetics, Cell Tropism, and Associated Immune Responses in SARS-CoV-2- and H5N1 Virus-Infected Human Induced Pluripotent Stem Cell-Derived Neural Models.mSphere. 2021 Jun 30;6(3):e0027021. doi: 10.1128/mSphere.00270-21. Epub 2021 Jun 23. mSphere. 2021. PMID: 34160239 Free PMC article.
-
Morphological, cellular, and molecular basis of brain infection in COVID-19 patients.Proc Natl Acad Sci U S A. 2022 Aug 30;119(35):e2200960119. doi: 10.1073/pnas.2200960119. Epub 2022 Aug 11. Proc Natl Acad Sci U S A. 2022. PMID: 35951647 Free PMC article.
-
Neurological manifestations of SARS-CoV-2: complexity, mechanism and associated disorders.Eur J Med Res. 2023 Aug 30;28(1):307. doi: 10.1186/s40001-023-01293-2. Eur J Med Res. 2023. PMID: 37649125 Free PMC article. Review.
-
A Review on SARS-CoV-2-Induced Neuroinflammation, Neurodevelopmental Complications, and Recent Updates on the Vaccine Development.Mol Neurobiol. 2021 Sep;58(9):4535-4563. doi: 10.1007/s12035-021-02399-6. Epub 2021 Jun 5. Mol Neurobiol. 2021. PMID: 34089508 Free PMC article. Review.
Cited by
-
Proteoform Analysis of the Human Olfactory System: A Window into Neurodegenerative Diseases.Proteomes. 2024 Mar 21;12(1):9. doi: 10.3390/proteomes12010009. Proteomes. 2024. PMID: 38535507 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

