Targeting the Structural Maturation Pathway of HIV-1 Reverse Transcriptase
- PMID: 38002285
- PMCID: PMC10669680
- DOI: 10.3390/biom13111603
Targeting the Structural Maturation Pathway of HIV-1 Reverse Transcriptase
Abstract
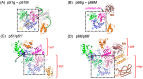
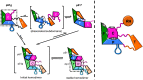
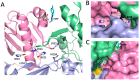
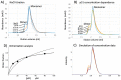
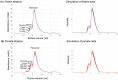
Formation of active HIV-1 reverse transcriptase (RT) proceeds via a structural maturation process that involves subdomain rearrangements and formation of an asymmetric p66/p66' homodimer. These studies were undertaken to evaluate whether the information about this maturation process can be used to identify small molecule ligands that retard or interfere with the steps involved. We utilized the isolated polymerase domain, p51, rather than p66, since the initial subdomain rearrangements are largely limited to this domain. Target sites at subdomain interfaces were identified and computational analysis used to obtain an initial set of ligands for screening. Chromatographic evaluations of the p51 homodimer/monomer ratio support the feasibility of this approach. Ligands that bind near the interfaces and a ligand that binds directly to a region of the fingers subdomain involved in subunit interface formation were identified, and the interactions were further characterized by NMR spectroscopy and X-ray crystallography. Although these ligands were found to reduce dimer formation, further efforts will be required to obtain ligands with higher binding affinity. In contrast with previous ligand identification studies performed on the RT heterodimer, subunit interface surfaces are solvent-accessible in the p51 and p66 monomers, making these constructs preferable for identification of ligands that directly interfere with dimerization.
Keywords: HIV-1 reverse transcriptase; RT dimerization inhibitor; RT polymerase domain; RT structural maturation; ground state stabilization; maturation inhibitors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
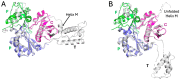

Similar articles
-
Homodimerization of the p51 subunit of HIV-1 reverse transcriptase.Biochemistry. 2010 Apr 6;49(13):2821-33. doi: 10.1021/bi902116z. Biochemistry. 2010. PMID: 20180596 Free PMC article.
-
Selective unfolding of one Ribonuclease H domain of HIV reverse transcriptase is linked to homodimer formation.Nucleic Acids Res. 2014 Apr;42(8):5361-77. doi: 10.1093/nar/gku143. Epub 2014 Feb 25. Nucleic Acids Res. 2014. PMID: 24574528 Free PMC article.
-
Proteolytic processing of an HIV-1 pol polyprotein precursor: insights into the mechanism of reverse transcriptase p66/p51 heterodimer formation.Int J Biochem Cell Biol. 2004 Sep;36(9):1836-47. doi: 10.1016/j.biocel.2004.02.020. Int J Biochem Cell Biol. 2004. PMID: 15183348
-
TSAO derivatives, inhibitors of HIV-1 reverse transcriptase dimerization: recent progress.Curr Pharm Des. 2006;12(15):1895-907. doi: 10.2174/138161206776873563. Curr Pharm Des. 2006. PMID: 16724955 Review.
-
Structural Maturation of HIV-1 Reverse Transcriptase-A Metamorphic Solution to Genomic Instability.Viruses. 2016 Sep 27;8(10):260. doi: 10.3390/v8100260. Viruses. 2016. PMID: 27690082 Free PMC article. Review.
Cited by
-
Microwave-assisted synthesis of a zirconium-based MOF as an efficient catalyst for one-pot synthesis of xanthene derivatives: in silico study as a potential anti-HIV RNA.RSC Adv. 2025 May 19;15(21):16654-16666. doi: 10.1039/d5ra02959g. eCollection 2025 May 15. RSC Adv. 2025. PMID: 40391364 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

