Elevated Levels of Mislocalised, Constitutive Ras Signalling Can Drive Quiescence by Uncoupling Cell-Cycle Regulation from Metabolic Homeostasis
- PMID: 38002301
- PMCID: PMC10669370
- DOI: 10.3390/biom13111619
Elevated Levels of Mislocalised, Constitutive Ras Signalling Can Drive Quiescence by Uncoupling Cell-Cycle Regulation from Metabolic Homeostasis
Abstract
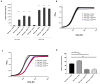
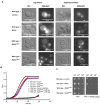
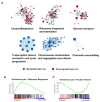
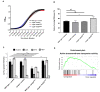
The small GTPase Ras plays an important role in connecting external and internal signalling cues to cell fate in eukaryotic cells. As such, the loss of RAS regulation, localisation, or expression level can drive changes in cell behaviour and fate. Post-translational modifications and expression levels are crucial to ensure Ras localisation, regulation, function, and cell fate, exemplified by RAS mutations and gene duplications that are common in many cancers. Here, we reveal that excessive production of yeast Ras2, in which the phosphorylation-regulated serine at position 225 is replaced with alanine or glutamate, leads to its mislocalisation and constitutive activation. Rather than inducing cell death, as has been widely reported to be a consequence of constitutive Ras2 signalling in yeast, the overexpression of RAS2S225A or RAS2S225E alleles leads to slow growth, a loss of respiration, reduced stress response, and a state of quiescence. These effects are mediated via cAMP/PKA signalling and transcriptional changes, suggesting that quiescence is promoted by an uncoupling of cell-cycle regulation from metabolic homeostasis. The quiescent cell fate induced by the overexpression of RAS2S225A or RAS2S225E could be rescued by the deletion of CUP9, a suppressor of the dipeptide transporter Ptr2, or the addition of peptone, implying that a loss of metabolic control, or a failure to pass a metabolic checkpoint, is central to this altered cell fate. Our data suggest that the combination of an increased RAS2 copy number and a dominant active mutation that leads to its mislocalisation can result in growth arrest and add weight to the possibility that approaches to retarget RAS signalling could be employed to develop new therapies.
Keywords: Ras; cell fate; metabolism; yeast signalling; yquiescence.
Conflict of interest statement
The authors declare that we have no conflicts of interest regarding this submission.
Figures


Similar articles
-
Regulation of respiratory growth by Ras: the glyoxylate cycle mutant, cit2Delta, is suppressed by RAS2.Curr Genet. 2006 Sep;50(3):161-71. doi: 10.1007/s00294-006-0084-z. Epub 2006 Jul 11. Curr Genet. 2006. PMID: 16832579
-
Whi2p links nutritional sensing to actin-dependent Ras-cAMP-PKA regulation and apoptosis in yeast.J Cell Sci. 2009 Mar 1;122(Pt 5):706-15. doi: 10.1242/jcs.042424. Epub 2009 Feb 10. J Cell Sci. 2009. PMID: 19208759 Free PMC article.
-
Ras2 and Ras1 protein phosphorylation in Saccharomyces cerevisiae.J Biol Chem. 1997 Jul 25;272(30):18790-800. doi: 10.1074/jbc.272.30.18790. J Biol Chem. 1997. PMID: 9228053
-
Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae.Mol Microbiol. 1999 Sep;33(5):904-18. doi: 10.1046/j.1365-2958.1999.01538.x. Mol Microbiol. 1999. PMID: 10476026 Review.
-
Metabolic control and gene dysregulation in yeast aging.Ann N Y Acad Sci. 2000 Jun;908:21-30. doi: 10.1111/j.1749-6632.2000.tb06632.x. Ann N Y Acad Sci. 2000. PMID: 10911944 Review.
Cited by
-
Editorial on the Research Topic 'Cell Differentiation, Oxidative Stress, and Oxygen Radicals-In Honor of Prof. Michael Breitenbach'.Biomolecules. 2024 Jul 29;14(8):920. doi: 10.3390/biom14080920. Biomolecules. 2024. PMID: 39199308 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

