Low-Entropy Hydration Shells at the Spike RBD's Binding Site May Reveal the Contagiousness of SARS-CoV-2 Variants
- PMID: 38002310
- PMCID: PMC10669249
- DOI: 10.3390/biom13111628
Low-Entropy Hydration Shells at the Spike RBD's Binding Site May Reveal the Contagiousness of SARS-CoV-2 Variants
Abstract
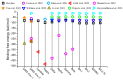
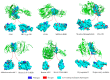
The infectivity of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is primarily determined by the binding affinity between the receptor-binding domain (RBD) of the spike protein and the angiotensin-converting enzyme 2 (ACE2) receptor. Here, through screening off pseudo hydrophilic groups on protein surfaces, the distribution of low-entropy regions on hydration shells of the ACE2 receptor and the RBDs of multiple SARS-CoV-2 variants was demonstrated. Shape matching between the low-entropy hydration shells of multiple SARS-CoV-2 variants and the ACE2 receptor has been identified as a mechanism that drives hydrophobic attraction between the RBDs and the ACE2 receptor, which estimates the binding affinity. Low-entropy regions of the hydration shells, which play important roles in determining the binding of other viruses and their receptors, are demonstrated. The RBD-ACE2 binding is thus found to be guided by hydrophobic collapse between the shape-matched low-entropy regions of the hydration shells of the proteins. A measure of the low-entropy status of the hydration shells can be estimated by calculating genuine hydrophilic groups within the binding sites. An important indicator of the contagiousness of SARS-CoV-2 variants is the low-entropy level of its hydration shells at the spike protein binding site.
Keywords: SARS-CoV-2 variants; contagiousness; low-entropy hydration shell; protein–protein interaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
SARS-CoV-2 Variants, RBD Mutations, Binding Affinity, and Antibody Escape.Int J Mol Sci. 2021 Nov 9;22(22):12114. doi: 10.3390/ijms222212114. Int J Mol Sci. 2021. PMID: 34829998 Free PMC article.
-
Sequence analysis of Indian SARS-CoV-2 isolates shows a stronger interaction of mutant receptor-binding domain with ACE2.Int J Infect Dis. 2021 Mar;104:491-500. doi: 10.1016/j.ijid.2021.01.020. Epub 2021 Jan 12. Int J Infect Dis. 2021. PMID: 33450373 Free PMC article.
-
Interactions of angiotensin-converting enzyme-2 (ACE2) and SARS-CoV-2 spike receptor-binding domain (RBD): a structural perspective.Mol Biol Rep. 2023 Mar;50(3):2713-2721. doi: 10.1007/s11033-022-08193-4. Epub 2022 Dec 23. Mol Biol Rep. 2023. PMID: 36562937 Free PMC article. Review.
-
The binding and structural basis of fox ACE2 to RBDs from different sarbecoviruses.Virol Sin. 2024 Aug;39(4):609-618. doi: 10.1016/j.virs.2024.06.004. Epub 2024 Jun 10. Virol Sin. 2024. PMID: 38866203 Free PMC article.
-
Structural basis of severe acute respiratory syndrome coronavirus 2 infection.Curr Opin HIV AIDS. 2021 Jan;16(1):74-81. doi: 10.1097/COH.0000000000000658. Curr Opin HIV AIDS. 2021. PMID: 33186231 Review.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

