Mitochondrial Proteomes in Neural Cells: A Systematic Review
- PMID: 38002320
- PMCID: PMC10669788
- DOI: 10.3390/biom13111638
Mitochondrial Proteomes in Neural Cells: A Systematic Review
Abstract
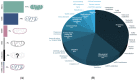
Mitochondria are ancient endosymbiotic double membrane organelles that support a wide range of eukaryotic cell functions through energy, metabolism, and cellular control. There are over 1000 known proteins that either reside within the mitochondria or are transiently associated with it. These mitochondrial proteins represent a functional subcellular protein network (mtProteome) that is encoded by mitochondrial and nuclear genomes and significantly varies between cell types and conditions. In neurons, the high metabolic demand and differential energy requirements at the synapses are met by specific modifications to the mtProteome, resulting in alterations in the expression and functional properties of the proteins involved in energy production and quality control, including fission and fusion. The composition of mtProteomes also impacts the localization of mitochondria in axons and dendrites with a growing number of neurodegenerative diseases associated with changes in mitochondrial proteins. This review summarizes the findings on the composition and properties of mtProteomes important for mitochondrial energy production, calcium and lipid signaling, and quality control in neural cells. We highlight strategies in mass spectrometry (MS) proteomic analysis of mtProteomes from cultured cells and tissue. The research into mtProteome composition and function provides opportunities in biomarker discovery and drug development for the treatment of metabolic and neurodegenerative disease.
Keywords: bioinformatics; energetics; evolution; mass spectrometry; neurodegeneration; protein quantification; synapses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Margulis L. Origin of Eukaryotic Cells: Evidence and Research Implications for a Theory of the Origin and Evolution of Microbial, Plant, and Animal Cells on the Precambrian Earth. Yale University Press; New Haven, CT, USA: 1970.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

