Gibberellic Acid Inhibits Dendrobium nobile- Piriformospora Symbiosis by Regulating the Expression of Cell Wall Metabolism Genes
- PMID: 38002331
- PMCID: PMC10669577
- DOI: 10.3390/biom13111649
Gibberellic Acid Inhibits Dendrobium nobile- Piriformospora Symbiosis by Regulating the Expression of Cell Wall Metabolism Genes
Abstract
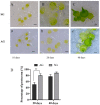
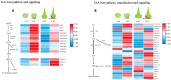
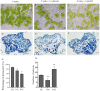
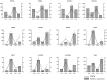
Orchid seeds lack endosperms and depend on mycorrhizal fungi for germination and nutrition acquisition under natural conditions. Piriformospora indica is a mycorrhizal fungus that promotes seed germination and seedling development in epiphytic orchids, such as Dendrobium nobile. To understand the impact of P. indica on D. nobile seed germination, we examined endogenous hormone levels by using liquid chromatography-mass spectrometry. We performed transcriptomic analysis of D. nobile protocorm at two developmental stages under asymbiotic germination (AG) and symbiotic germination (SG) conditions. The result showed that the level of endogenous IAA in the SG protocorm treatments was significantly higher than that in the AG protocorm treatments. Meanwhile, GA3 was only detected in the SG protocorm stages. IAA and GA synthesis and signaling genes were upregulated in the SG protocorm stages. Exogenous GA3 application inhibited fungal colonization inside the protocorm, and a GA biosynthesis inhibitor (PAC) promoted fungal colonization. Furthermore, we found that PAC prevented fungal hyphae collapse and degeneration in the protocorm, and differentially expressed genes related to cell wall metabolism were identified between the SG and AG protocorm stages. Exogenous GA3 upregulated SRC2 and LRX4 expression, leading to decreased fungal colonization. Meanwhile, GA inhibitors upregulated EXP6, EXB16, and EXP10-2 expression, leading to increased fungal colonization. Our findings suggest that GA regulates the expression of cell wall metabolism genes in D. nobile, thereby inhibiting the establishment of mycorrhizal symbiosis.
Keywords: cell wall; hormones; protocorm; symbiosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
What role does the seed coat play during symbiotic seed germination in orchids: an experimental approach with Dendrobium officinale.BMC Plant Biol. 2022 Jul 29;22(1):375. doi: 10.1186/s12870-022-03760-0. BMC Plant Biol. 2022. PMID: 35906552 Free PMC article.
-
Symbiotic and Asymbiotic Germination of Dendrobium officinale (Orchidaceae) Respond Differently to Exogenous Gibberellins.Int J Mol Sci. 2020 Aug 25;21(17):6104. doi: 10.3390/ijms21176104. Int J Mol Sci. 2020. PMID: 32854186 Free PMC article.
-
Fungi isolated from host protocorms accelerate symbiotic seed germination in an endangered orchid species (Dendrobium chrysotoxum) from southern China.Mycorrhiza. 2020 Jul;30(4):529-539. doi: 10.1007/s00572-020-00964-w. Epub 2020 Jun 19. Mycorrhiza. 2020. PMID: 32562087
-
A perspective on orchid seed and protocorm development.Bot Stud. 2017 Dec;58(1):33. doi: 10.1186/s40529-017-0188-4. Epub 2017 Aug 4. Bot Stud. 2017. PMID: 28779349 Free PMC article. Review.
-
Symbiotic in vitro seed propagation of Dendrobium: fungal and bacterial partners and their influence on plant growth and development.Planta. 2015 Jul;242(1):1-22. doi: 10.1007/s00425-015-2301-9. Epub 2015 May 5. Planta. 2015. PMID: 25940846 Review.
Cited by
-
Excessive accumulation of auxin inhibits protocorm development during germination of Paphiopedilum spicerianum.Plant Cell Rep. 2025 Jan 6;44(1):23. doi: 10.1007/s00299-024-03419-0. Plant Cell Rep. 2025. PMID: 39762613
References
-
- Fochi V., Chitarra W., Kohler A., Voyron S., Singan V.R., Lindquist E.A., Barry K.W., Girlanda M., Grigoriev I.V., Martin F., et al. Fungal and plant gene expression in the Tulasnella calospora-Serapias vomeracea symbiosis provides clues about nitrogen pathways in orchid mycorrhizas. New Phytol. 2017;213:365–379. doi: 10.1111/nph.14279. - DOI - PubMed
-
- Rasmussen H.N. Cell-differentiation and mycorrhizal infection in Dactylorhiza-majalis (Rchb. f.) Hunt & Summerh (orchidaceae) during germination in vitro. New Phytol. 1990;116:137–147.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

