LEO1 Is Required for Efficient Entry into Quiescence, Control of H3K9 Methylation and Gene Expression in Human Fibroblasts
- PMID: 38002344
- PMCID: PMC10668985
- DOI: 10.3390/biom13111662
LEO1 Is Required for Efficient Entry into Quiescence, Control of H3K9 Methylation and Gene Expression in Human Fibroblasts
Abstract
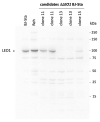
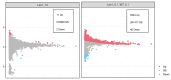
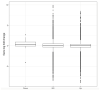
(1) Background: The LEO1 (Left open reading frame 1) protein is a conserved subunit of the PAF1C complex (RNA polymerase II-associated factor 1 complex). PAF1C has well-established mechanistic functions in elongation of transcription and RNA processing. We previously showed, in fission yeast, that LEO1 controls histone H3K9 methylation levels by affecting the turnover of histone H3 in chromatin, and that it is essential for the proper regulation of gene expression during cellular quiescence. Human fibroblasts enter a reversible quiescence state upon serum deprivation in the growth media. Here we investigate the function of LEO1 in human fibroblasts. (2) Methods: We knocked out the LEO1 gene using CRISPR/Cas9 methodology in human fibroblasts and verified that the LEO1 protein was undetectable by Western blot. We characterized the phenotype of the ΔLEO1 knockout cells with FACS analysis and cell growth assays. We used RNA-sequencing using spike-in controls to measure gene expression and spike-in controlled ChIP-sequencing experiments to measure the histone modification H3K9me2 genome-wide. (3) Results: Gene expression levels are altered in quiescent cells, however factors controlling chromatin and gene expression changes in quiescent human cells are largely unknown. The ΔLEO1 knockout fibroblasts are viable but have reduced metabolic activity compared to wild-type cells. ΔLEO1 cells showed a slower entry into quiescence and a different morphology compared to wild-type cells. Gene expression was generally reduced in quiescent wild-type cells. The downregulated genes included genes involved in cell proliferation. A small number of genes were upregulated in quiescent wild-type cells including several genes involved in ERK1/ERK2 and Wnt signaling. In quiescent ΔLEO1 cells, many genes were mis-regulated compared to wild-type cells. This included genes involved in Calcium ion transport and cell morphogenesis. Finally, spike-in controlled ChIP-sequencing experiments demonstrated that the histone modification H3K9me2 levels are globally increased in quiescent ΔLEO1 cells. (4) Conclusions: Thus, LEO1 is important for proper entry into cellular quiescence, control of H3K9me2 levels, and gene expression in human fibroblasts.
Keywords: H3K9me2; LEO1; PAF1C; cellular quiescence; chromatin; fibroblasts; gene expression; histone modification.
Conflict of interest statement
The authors have no conflict of interests and no competing interest.
Figures



Similar articles
-
Leo1 is essential for the dynamic regulation of heterochromatin and gene expression during cellular quiescence.Epigenetics Chromatin. 2019 Jul 17;12(1):45. doi: 10.1186/s13072-019-0292-7. Epigenetics Chromatin. 2019. PMID: 31315658 Free PMC article.
-
The Paf1 complex factors Leo1 and Paf1 promote local histone turnover to modulate chromatin states in fission yeast.EMBO Rep. 2015 Dec;16(12):1673-87. doi: 10.15252/embr.201541214. Epub 2015 Oct 29. EMBO Rep. 2015. PMID: 26518661 Free PMC article.
-
An essential role for the Ino80 chromatin remodeling complex in regulation of gene expression during cellular quiescence.Chromosome Res. 2023 Apr 12;31(2):14. doi: 10.1007/s10577-023-09723-x. Chromosome Res. 2023. PMID: 37043046 Free PMC article.
-
Regulation of the SUV39H Family Methyltransferases: Insights from Fission Yeast.Biomolecules. 2023 Mar 25;13(4):593. doi: 10.3390/biom13040593. Biomolecules. 2023. PMID: 37189341 Free PMC article. Review.
-
Epigenomics in stress tolerance of plants under the climate change.Mol Biol Rep. 2023 Jul;50(7):6201-6216. doi: 10.1007/s11033-023-08539-6. Epub 2023 Jun 9. Mol Biol Rep. 2023. PMID: 37294468 Review.
Cited by
-
The Wild West of spike-in normalization.Nat Biotechnol. 2024 Sep;42(9):1343-1349. doi: 10.1038/s41587-024-02377-y. Nat Biotechnol. 2024. PMID: 39271835 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

