The Molecular Basis for Selectivity of the Cytotoxic Response of Lung Adenocarcinoma Cells to Cold Atmospheric Plasma
- PMID: 38002354
- PMCID: PMC10669024
- DOI: 10.3390/biom13111672
The Molecular Basis for Selectivity of the Cytotoxic Response of Lung Adenocarcinoma Cells to Cold Atmospheric Plasma
Abstract
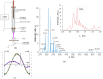
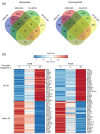
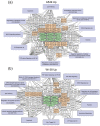
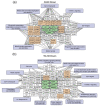
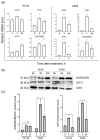
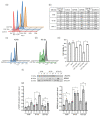
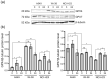
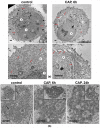
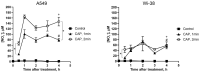
The interaction of cold atmospheric plasma (CAP) with biotargets is accompanied by chemical reactions on their surfaces and insides, and it has great potential as an anticancer approach. This study discovers the molecular mechanisms that may explain the selective death of tumor cells under CAP exposure. To reach this goal, the transcriptional response to CAP treatment was analyzed in A549 lung adenocarcinoma cells and in lung-fibroblast Wi-38 cells. We found that the CAP treatment induced the common trend of response from A549 and Wi-38 cells-the p53 pathway, KRAS signaling, UV response, TNF-alpha signaling, and apoptosis-related processes were up-regulated in both cell lines. However, the amplitude of the response to CAP was more variable in the A549 cells. The CAP-dependent death of A549 cells was accompanied by DNA damage, cell-cycle arrest in G2/M, and the dysfunctional response of glutathione peroxidase 4 (GPx4). The activation of the genes of endoplasmic reticulum stress and ER lumens was detected only in the A549 cells. Transmission-electron microscopy confirmed the alteration of the morphology of the ER lumens in the A549 cells after the CAP exposure. It can be concluded that the responses to nuclear stress and ER stress constitute the main differences in the sensitivity of tumor and healthy cells to CAP exposure.
Keywords: GADD45; GPx4; cell-cycle arrest; cold atmospheric plasma; endoplasmic reticulum stress; p53 pathway; transcriptome analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Apoptotic effects of cold atmospheric pressure plasma on A549 and LL/2 lung carcinoma cell lines.Sci Rep. 2025 Jun 4;15(1):19567. doi: 10.1038/s41598-025-03934-4. Sci Rep. 2025. PMID: 40467678 Free PMC article.
-
Chloroquine Enhances Death in Lung Adenocarcinoma A549 Cells Exposed to Cold Atmospheric Plasma Jet.Cells. 2023 Jan 12;12(2):290. doi: 10.3390/cells12020290. Cells. 2023. PMID: 36672225 Free PMC article.
-
The antiproliferative effects of cold atmospheric plasma-activated media on different cancer cell lines, the implication of ozone as a possible underlying mechanism.J Cell Physiol. 2019 May;234(5):6778-6782. doi: 10.1002/jcp.27428. Epub 2018 Nov 1. J Cell Physiol. 2019. PMID: 30387137
-
Intracellular Responses Triggered by Cold Atmospheric Plasma and Plasma-Activated Media in Cancer Cells.Molecules. 2021 Mar 2;26(5):1336. doi: 10.3390/molecules26051336. Molecules. 2021. PMID: 33801451 Free PMC article. Review.
-
Toward understanding the selective anticancer capacity of cold atmospheric plasma--a model based on aquaporins (Review).Biointerphases. 2015 Dec 22;10(4):040801. doi: 10.1116/1.4938020. Biointerphases. 2015. PMID: 26700469 Review.
Cited by
-
Bacterial Decontamination of Water-Containing Objects Using Piezoelectric Direct Discharge Plasma and Plasma Jet.Biomolecules. 2024 Feb 2;14(2):181. doi: 10.3390/biom14020181. Biomolecules. 2024. PMID: 38397418 Free PMC article.
-
Apoptotic effects of cold atmospheric pressure plasma on A549 and LL/2 lung carcinoma cell lines.Sci Rep. 2025 Jun 4;15(1):19567. doi: 10.1038/s41598-025-03934-4. Sci Rep. 2025. PMID: 40467678 Free PMC article.
-
Dissecting the METTL3/STC2 axis in colorectal cancer: implications for drug resistance and metastasis.Cell Biol Toxicol. 2025 Jun 10;41(1):100. doi: 10.1007/s10565-025-10043-5. Cell Biol Toxicol. 2025. PMID: 40494964 Free PMC article.
References
-
- Patrakova E., Biryukov M., Troitskaya O., Gugin P., Milakhina E., Semenov D., Poletaeva J., Ryabchikova E., Novak D., Kryachkova N., et al. Chloroquine Enhances Death in Lung Adenocarcinoma A549 Cells Exposed to Cold Atmospheric Plasma Jet. Cells. 2023;12:290. doi: 10.3390/cells12020290. - DOI - PMC - PubMed
-
- Schweigert I., Zakrevsky D., Gugin P., Yelak E., Golubitskaya E., Troitskaya O., Koval O. Interaction of Cold Atmospheric Argon and Helium Plasma Jets with Bio-Target with Grounded Substrate Beneath. Appl. Sci. 2019;9:4528. doi: 10.3390/app9214528. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

