Multivariable Risk Modelling and Survival Analysis with Machine Learning in SARS-CoV-2 Infection
- PMID: 38002776
- PMCID: PMC10672177
- DOI: 10.3390/jcm12227164
Multivariable Risk Modelling and Survival Analysis with Machine Learning in SARS-CoV-2 Infection
Abstract
Aim: To evaluate the performance of a machine learning model based on demographic variables, blood tests, pre-existing comorbidities, and computed tomography(CT)-based radiomic features to predict critical outcome in patients with acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Methods: We retrospectively enrolled 694 SARS-CoV-2-positive patients. Clinical and demographic data were extracted from clinical records. Radiomic data were extracted from CT. Patients were randomized to the training (80%, n = 556) or test (20%, n = 138) dataset. The training set was used to define the association between severity of disease and comorbidities, laboratory tests, demographic, and CT-based radiomic variables, and to implement a risk-prediction model. The model was evaluated using the C statistic and Brier scores. The test set was used to assess model prediction performance.
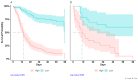
Results: Patients who died (n = 157) were predominantly male (66%) over the age of 50 with median (range) C-reactive protein (CRP) = 5 [1, 37] mg/dL, lactate dehydrogenase (LDH) = 494 [141, 3631] U/I, and D-dimer = 6.006 [168, 152.015] ng/mL. Surviving patients (n = 537) had median (range) CRP = 3 [0, 27] mg/dL, LDH = 484 [78, 3.745] U/I, and D-dimer = 1.133 [96, 55.660] ng/mL. The strongest risk factors were D-dimer, age, and cardiovascular disease. The model implemented using the variables identified using the LASSO Cox regression analysis classified 90% of non-survivors as high-risk individuals in the testing dataset. In this sample, the estimated median survival in the high-risk group was 9 days (95% CI; 9-37), while the low-risk group did not reach the median survival of 50% (p < 0.001).
Conclusions: A machine learning model based on combined data available on the first days of hospitalization (demographics, CT-radiomics, comorbidities, and blood biomarkers), can identify SARS-CoV-2 patients at risk of serious illness and death.
Keywords: CT; SARS-CoV-2; machine learning; radiomics; survival.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
High serum lactate dehydrogenase and dyspnea: Positive predictors of adverse outcome in critical COVID-19 patients in Yichang.World J Clin Cases. 2020 Nov 26;8(22):5535-5546. doi: 10.12998/wjcc.v8.i22.5535. World J Clin Cases. 2020. PMID: 33344544 Free PMC article.
-
Machine Learning-Based Prediction of COVID-19 Severity and Progression to Critical Illness Using CT Imaging and Clinical Data.Korean J Radiol. 2021 Jul;22(7):1213-1224. doi: 10.3348/kjr.2020.1104. Epub 2021 Mar 9. Korean J Radiol. 2021. PMID: 33739635 Free PMC article.
-
Machine learning-based prognostic modeling using clinical data and quantitative radiomic features from chest CT images in COVID-19 patients.Comput Biol Med. 2021 May;132:104304. doi: 10.1016/j.compbiomed.2021.104304. Epub 2021 Mar 3. Comput Biol Med. 2021. PMID: 33691201 Free PMC article.
-
[Clinical features and risk factors for secondary hemophagocytic lymphohistiocytosis in elderly patients with severe SARS-CoV-2 infection: a multicenter retrospective cohort study].Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2023 Aug;35(8):793-799. doi: 10.3760/cma.j.cn121430-20230510-00158. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2023. PMID: 37593855 Chinese.
-
Evaluation of novel coronavirus disease (COVID-19) using quantitative lung CT and clinical data: prediction of short-term outcome.Eur Radiol Exp. 2020 Jun 26;4(1):39. doi: 10.1186/s41747-020-00167-0. Eur Radiol Exp. 2020. PMID: 32592118 Free PMC article.
Cited by
-
Does FDG PET-Based Radiomics Have an Added Value for Prediction of Overall Survival in Non-Small Cell Lung Cancer?J Clin Med. 2024 Apr 29;13(9):2613. doi: 10.3390/jcm13092613. J Clin Med. 2024. PMID: 38731142 Free PMC article.
-
Enhancing the Understanding of Abdominal Trauma During the COVID-19 Pandemic Through Co-Occurrence Analysis and Machine Learning.Diagnostics (Basel). 2024 Oct 31;14(21):2444. doi: 10.3390/diagnostics14212444. Diagnostics (Basel). 2024. PMID: 39518411 Free PMC article.
References
-
- Lambrou A.S., Shirk P., Steele M.K., Paul P., Paden C.R., Cadwell B., Reese H.E., Aoki Y., Hassell N., Zheng X.Y., et al. Genomic surveillance for SARS-CoV-2 variants: Predominance of the delta (b.1.617.2) and omicron (b.1.1.529) variants—United states, June 2021–January 2022. Morb. Mortal. Wkly. Rep. 2022;71:206–211. doi: 10.15585/mmwr.mm7106a4. - DOI - PMC - PubMed
-
- Colson P., Delerce J., Burel E., Dahan J., Jouffret A., Fenollar F., Yahi N., Fantini J., La Scola B., Raoult D. Emergence in southern france of a new SARS-CoV-2 variant harbouring both n501y and e484k substitutions in the spike protein. Arch. Virol. 2022;167:1185–1190. doi: 10.1007/s00705-022-05385-y. - DOI - PMC - PubMed
-
- Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., Northwell COVID-19 Research Consortium Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

