Finding Predictors of Leg Defects in Pigs Using CNV-GWAS
- PMID: 38002997
- PMCID: PMC10671522
- DOI: 10.3390/genes14112054
Finding Predictors of Leg Defects in Pigs Using CNV-GWAS
Abstract
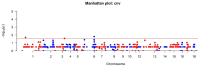
One of the most important areas of modern genome research is the search for meaningful relationships between genetic variants and phenotypes. In the livestock field, there has been research demonstrating the influence of copy number variants (CNVs) on phenotypic variation. Despite the wide range in the number and size of detected CNVs, a significant proportion differ between breeds and their functional effects are underestimated in the pig industry. In this work, we focused on the problem of leg defects in pigs (lumps/growths in the area of the hock joint on the hind legs) and focused on searching for molecular genetic predictors associated with this trait for the selection of breeding stock. The study was conducted on Large White pigs using three CNV calling tools (PennCNV, QuantiSNP and R-GADA) and the CNVRanger association analysis tool (CNV-GWAS). As a result, the analysis identified three candidate CNVRs associated with the formation of limb defects. Subsequent functional analysis suggested that all identified CNVs may act as potential predictors of the hock joint phenotype of pigs. It should be noted that the results obtained indicate that all significant regions are localized in genes (CTH, SRSF11, MAN1A1 and LPIN1) responsible for the metabolism of amino acids, fatty acids, glycerolipids and glycerophospholipids, thereby related to the immune response, liver functions, content intramuscular fat and animal fatness. These results are consistent with previously published studies, according to which a predisposition to the formation of leg defects can be realized through genetic variants associated with the functions of the liver, kidneys and hematological characteristics.
Keywords: CNV; GWAS; SNP; leg defects; pig.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
A comprehensive survey of copy number variation in 18 diverse pig populations and identification of candidate copy number variable genes associated with complex traits.BMC Genomics. 2012 Dec 27;13:733. doi: 10.1186/1471-2164-13-733. BMC Genomics. 2012. PMID: 23270433 Free PMC article.
-
Genome-wide detection of CNV regions and their potential association with growth and fatness traits in Duroc pigs.BMC Genomics. 2021 May 8;22(1):332. doi: 10.1186/s12864-021-07654-7. BMC Genomics. 2021. PMID: 33964879 Free PMC article.
-
A composite strategy of genome-wide association study and copy number variation analysis for carcass traits in a Duroc pig population.BMC Genomics. 2022 Aug 13;23(1):590. doi: 10.1186/s12864-022-08804-1. BMC Genomics. 2022. PMID: 35964005 Free PMC article.
-
Analysis of genome-wide copy number variations in Chinese indigenous and western pig breeds by 60 K SNP genotyping arrays.PLoS One. 2014 Sep 8;9(9):e106780. doi: 10.1371/journal.pone.0106780. eCollection 2014. PLoS One. 2014. PMID: 25198154 Free PMC article.
-
Genome wide copy number variations using Porcine 60K SNP Beadchip in Landlly pigs.Anim Biotechnol. 2023 Nov;34(6):1891-1899. doi: 10.1080/10495398.2022.2056047. Epub 2022 Apr 4. Anim Biotechnol. 2023. PMID: 35369845 Review.
Cited by
-
Genomic and transcriptomic analyses reveal the genetic basis of leg diseases in laying hens.Poult Sci. 2025 Mar;104(3):104887. doi: 10.1016/j.psj.2025.104887. Epub 2025 Feb 13. Poult Sci. 2025. PMID: 39970519 Free PMC article.
-
Identifying Significant SNPs of the Total Number of Piglets Born and Their Relationship with Leg Bumps in Pigs.Biology (Basel). 2024 Dec 11;13(12):1034. doi: 10.3390/biology13121034. Biology (Basel). 2024. PMID: 39765701 Free PMC article.
-
Visualization of Runs of Homozygosity and Classification Using Convolutional Neural Networks.Biology (Basel). 2025 Apr 16;14(4):426. doi: 10.3390/biology14040426. Biology (Basel). 2025. PMID: 40282291 Free PMC article.
-
Genetic Architecture of Hock Joint Bumps in Pigs: Insights from ROH and GWAS Analyses.Animals (Basel). 2025 Apr 20;15(8):1178. doi: 10.3390/ani15081178. Animals (Basel). 2025. PMID: 40282012 Free PMC article.
References
-
- Wright D., Boije H., Meadows J.R., Bed’hom B., Gourichon D., Vieaud A., Tixier-Boichard M., Rubin C.J., Imsland F., Hallböök F., et al. Copy number variation in intron 1 of SOX5 causes the Pea-comb phenotype in chickens. PLoS Genet. 2009;5:e1000512. doi: 10.1371/journal.pgen.1000512. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials