Identification and Functional Characterization of Two Major Loci Associated with Resistance against Brown Planthoppers (Nilaparvata lugens (Stål)) Derived from Oryza nivara
- PMID: 38003009
- PMCID: PMC10671472
- DOI: 10.3390/genes14112066
Identification and Functional Characterization of Two Major Loci Associated with Resistance against Brown Planthoppers (Nilaparvata lugens (Stål)) Derived from Oryza nivara
Abstract
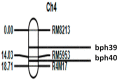
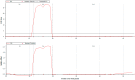
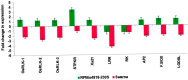
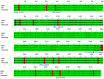
The brown planthopper (BPH) is a highly destructive pest of rice, causing significant economic losses in various regions of South and Southeast Asia. Researchers have made promising strides in developing resistance against BPH in rice. Introgression line RPBio4918-230S, derived from Oryza nivara, has shown consistent resistance to BPH at both the seedling and adult stages of rice plants. Segregation analysis has revealed that this resistance is governed by two recessive loci, known as bph39(t) and bph40(t), contributing to 21% and 22% of the phenotypic variance, respectively. We later mapped the genes using a backcross population derived from a cross between Swarna and RPBio4918-230S. We identified specific marker loci, namely RM8213, RM5953, and R4M17, on chromosome 4, flanking the bph39(t) and bph40(t) loci. Furthermore, quantitative expression analysis of candidate genes situated between the RM8213 and R4M17 markers was conducted. It was observed that eight genes exhibited up-regulation in RPBio4918-230S and down-regulation in Swarna after BPH infestation. One gene of particular interest, a serine/threonine-protein kinase receptor (STPKR), showed significant up-regulation in RPBio4918-230S. In-depth sequencing of the susceptible and resistant alleles of STPKR from Swarna and RPBio4918-230S, respectively, revealed numerous single nucleotide polymorphisms (SNPs) and insertion-deletion (InDel) mutations, both in the coding and regulatory regions of the gene. Notably, six of these mutations resulted in amino acid substitutions in the coding region of STPKR (R5K, I38L, S120N, T319A, T320S, and F348S) when compared to Swarna and the reference sequence of Nipponbare. Further validation of these mutations in a set of highly resistant and susceptible backcross inbred lines confirmed the candidacy of the STPKR gene with respect to BPH resistance controlled by bph39(t) and bph40(t). Functional markers specific for STPKR have been developed and validated and can be used for accelerated transfer of the resistant locus to elite rice cultivars.
Keywords: Oryza nivara; STPKR gene; brown planthopper (BPH); rice.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Genetics of novel brown planthopper Nilaparvata lugens (Stål) resistance genes in derived introgression lines from the interspecific cross O. sativa var. Swarna × O. nivara.J Genet. 2019 Dec;98:113. J Genet. 2019. PMID: 31819024
-
High-resolution genetic mapping of a novel brown planthopper resistance locus, Bph34 in Oryza sativa L. X Oryza nivara (Sharma & Shastry) derived interspecific F2 population.Theor Appl Genet. 2018 May;131(5):1163-1171. doi: 10.1007/s00122-018-3069-7. Epub 2018 Feb 23. Theor Appl Genet. 2018. PMID: 29476225
-
Molecular mapping and transfer of a novel brown planthopper resistance gene bph42 from Oryza rufipogon (Griff.) To cultivated rice (Oryza sativa L.).Mol Biol Rep. 2022 Sep;49(9):8597-8606. doi: 10.1007/s11033-022-07692-8. Epub 2022 Jun 28. Mol Biol Rep. 2022. PMID: 35764746
-
Recent Advances in the Genetic and Biochemical Mechanisms of Rice Resistance to Brown Planthoppers (Nilaparvata lugens Stål).Int J Mol Sci. 2023 Nov 30;24(23):16959. doi: 10.3390/ijms242316959. Int J Mol Sci. 2023. PMID: 38069282 Free PMC article. Review.
-
Genetic and biochemical mechanisms of rice resistance to planthopper.Plant Cell Rep. 2016 Aug;35(8):1559-72. doi: 10.1007/s00299-016-1962-6. Epub 2016 Mar 15. Plant Cell Rep. 2016. PMID: 26979747 Review.
Cited by
-
Breeding for brown plant hopper resistance in rice: recent updates and future perspectives.Mol Biol Rep. 2024 Oct 4;51(1):1038. doi: 10.1007/s11033-024-09966-9. Mol Biol Rep. 2024. PMID: 39365503 Review.
-
Recent Advances in Gene Mining and Hormonal Mechanism for Brown Planthopper Resistance in Rice.Int J Mol Sci. 2024 Dec 2;25(23):12965. doi: 10.3390/ijms252312965. Int J Mol Sci. 2024. PMID: 39684676 Free PMC article. Review.
References
-
- Chen C.N., Cheng C.C. The population levels of Nilaparvata lugens (Stål) in relation to the yield loss of rice. Plant Prot. Bull. 1978;20:197–209.
-
- Heong K.L. Are planthopper problems due to breakdown in ecosystem services? In: Heong K.L., Hardy B., editors. Plant Hoppers—New Threats to the Sustainability of Intensive Rice Production Systems in Asia. International Rice Research Institute; Los Banos, Philippines: 2009. pp. 221–232.
-
- Zhu P., Zheng X., Xu H., Johnson A.C., Heong K.L., Gurr G.M., Lu Z. Nitrogen fertilization of rice plants improves ecological fitness of an entomophagous predator but dampens its impact on prey, the rice brown planthopper, Nilaparvata lugens. J. Pest Sci. 2020;93:747–755. doi: 10.1007/s10340-019-01174-w. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

