Genome-Wide Identification and Preliminary Functional Analysis of BAM (β-Amylase) Gene Family in Upland Cotton
- PMID: 38003020
- PMCID: PMC10671626
- DOI: 10.3390/genes14112077
Genome-Wide Identification and Preliminary Functional Analysis of BAM (β-Amylase) Gene Family in Upland Cotton
Abstract
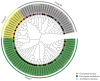
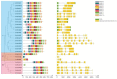
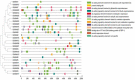
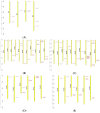
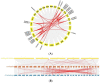
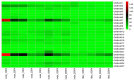
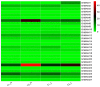
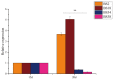
The β-amylase (BAM) gene family encodes important enzymes that catalyze the conversion of starch to maltose in various biological processes of plants and play essential roles in regulating the growth and development of multiple plants. So far, BAMs have been extensively studied in Arabidopsis thaliana (A. thaliana). However, the characteristics of the BAM gene family in the crucial economic crop, cotton, have not been reported. In this study, 27 GhBAM genes in the genome of Gossypium hirsutum L (G. hirsutum) were identified by genome-wide identification, and they were divided into three groups according to sequence similarity and phylogenetic relationship. The gene structure, chromosome distribution, and collinearity of all GhBAM genes identified in the genome of G. hirsutum were analyzed. Further sequence alignment of the core domain of glucosyl hydrolase showed that all GhBAM family genes had the glycosyl hydrolase family 14 domain. We identified the BAM gene GhBAM7 and preliminarily investigated its function by transcriptional sequencing analysis, qRT-PCR, and subcellular localization. These results suggested that the GhBAM7 gene may influence fiber strength during fiber development. This systematic analysis provides new insight into the transcriptional characteristics of BAM genes in G. hirsutum. It may lay the foundation for further study of the function of these genes.
Keywords: fiber development; genome-wide identification; starch metabolism; upland cotton; β-amylase (BAM) gene family.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Genome-wide cloning, identification, classification and functional analysis of cotton heat shock transcription factors in cotton (Gossypium hirsutum).BMC Genomics. 2014 Nov 6;15(1):961. doi: 10.1186/1471-2164-15-961. BMC Genomics. 2014. PMID: 25378022 Free PMC article.
-
Genome-wide identification of BAM (β-amylase) gene family in jujube (Ziziphus jujuba Mill.) and expression in response to abiotic stress.BMC Genomics. 2022 Jun 13;23(1):438. doi: 10.1186/s12864-022-08630-5. BMC Genomics. 2022. PMID: 35698031 Free PMC article.
-
Genome-Wide Analysis of the NF-YB Gene Family in Gossypium hirsutum L. and Characterization of the Role of GhDNF-YB22 in Embryogenesis.Int J Mol Sci. 2018 Feb 6;19(2):483. doi: 10.3390/ijms19020483. Int J Mol Sci. 2018. PMID: 29415481 Free PMC article.
-
Genome-wide identification and characterization of SnRK2 gene family in cotton (Gossypium hirsutum L.).BMC Genet. 2017 Jun 12;18(1):54. doi: 10.1186/s12863-017-0517-3. BMC Genet. 2017. PMID: 28606097 Free PMC article.
-
The evolution of functional complexity within the β-amylase gene family in land plants.BMC Evol Biol. 2019 Feb 28;19(1):66. doi: 10.1186/s12862-019-1395-2. BMC Evol Biol. 2019. PMID: 30819112 Free PMC article.
Cited by
-
Transcriptome Analysis Deciphers the Underlying Molecular Mechanism of Peanut Lateral Branch Angle Formation Using Erect Branching Mutant.Genes (Basel). 2024 Oct 21;15(10):1348. doi: 10.3390/genes15101348. Genes (Basel). 2024. PMID: 39457471 Free PMC article.
-
Genome-Wide and Expression Pattern Analysis of the HIT4 Gene Family Uncovers the Involvement of GHHIT4_4 in Response to Verticillium Wilt in Gossypium hirsutum.Genes (Basel). 2024 Mar 9;15(3):348. doi: 10.3390/genes15030348. Genes (Basel). 2024. PMID: 38540407 Free PMC article.
-
A Genome-Wide Analysis of the BAM Gene Family and Identification of the Cold-Responsive Genes in Pomegranate (Punica granatum L.).Plants (Basel). 2024 May 10;13(10):1321. doi: 10.3390/plants13101321. Plants (Basel). 2024. PMID: 38794392 Free PMC article.
-
Genome-Wide Identification and Expression Profiling of the α-Amylase (AMY) Gene Family in Potato.Genes (Basel). 2024 Jun 17;15(6):793. doi: 10.3390/genes15060793. Genes (Basel). 2024. PMID: 38927729 Free PMC article.
-
Genome-wide analysis of the FKBP gene family and the potential role of GhFKBP 13 in chloroplast biogenesis in upland cotton.BMC Genomics. 2025 Feb 10;26(1):125. doi: 10.1186/s12864-025-11293-7. BMC Genomics. 2025. PMID: 39930370 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

