Comprehensive Identification of Mitochondrial Pseudogenes (NUMTs) in the Human Telomere-to-Telomere Reference Genome
- PMID: 38003036
- PMCID: PMC10671835
- DOI: 10.3390/genes14112092
Comprehensive Identification of Mitochondrial Pseudogenes (NUMTs) in the Human Telomere-to-Telomere Reference Genome
Abstract
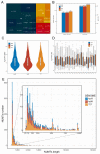
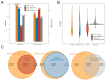
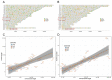
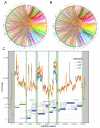
Practices related to mitochondrial research have long been hindered by the presence of mitochondrial pseudogenes within the nuclear genome (NUMTs). Even though partially assembled human reference genomes like hg38 have included NUMTs compilation, the exhaustive NUMTs within the only complete reference genome (T2T-CHR13) remain unknown. Here, we comprehensively identified the fixed NUMTs within the reference genome using human pan-mitogenome (HPMT) from GeneBank. The inclusion of HPMT serves the purpose of establishing an authentic mitochondrial DNA (mtDNA) mutational spectrum for the identification of NUMTs, distinguishing it from the polymorphic variations found in NUMTs. Using HPMT, we identified approximately 10% of additional NUMTs in three human reference genomes under stricter thresholds. And we also observed an approximate 6% increase in NUMTs in T2T-CHR13 compared to hg38, including NUMTs on the short arms of chromosomes 13, 14, and 15 that were not assembled previously. Furthermore, alignments based on 20-mer from mtDNA suggested the presence of more mtDNA-like short segments within the nuclear genome, which should be avoided for short amplicon or cell free mtDNA detection. Finally, through the assay of transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) on cell lines before and after mtDNA elimination, we concluded that NUMTs have a minimal impact on bulk ATAC-seq, even though 16% of sequencing data originated from mtDNA.
Keywords: ATAC-seq; NUMTs; human reference genome; mitochondrial DNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
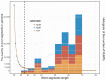
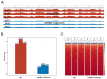
Similar articles
-
Mitochondrial pseudogenes in the nuclear genome of Aedes aegypti mosquitoes: implications for past and future population genetic studies.BMC Genet. 2009 Mar 6;10:11. doi: 10.1186/1471-2156-10-11. BMC Genet. 2009. PMID: 19267896 Free PMC article.
-
Low number of mitochondrial pseudogenes in the chicken (Gallus gallus) nuclear genome: implications for molecular inference of population history and phylogenetics.BMC Evol Biol. 2004 Jun 25;4:17. doi: 10.1186/1471-2148-4-17. BMC Evol Biol. 2004. PMID: 15219233 Free PMC article.
-
Distribution of nuclear mitochondrial DNA in cattle nuclear genome.J Anim Breed Genet. 2007 Oct;124(5):264-8. doi: 10.1111/j.1439-0388.2007.00674.x. J Anim Breed Genet. 2007. PMID: 17868078
-
The Mighty NUMT: Mitochondrial DNA Flexing Its Code in the Nuclear Genome.Biomolecules. 2023 Apr 27;13(5):753. doi: 10.3390/biom13050753. Biomolecules. 2023. PMID: 37238623 Free PMC article. Review.
-
Interpreting NUMTs in forensic genetics: Seeing the forest for the trees.Forensic Sci Int Genet. 2021 Jul;53:102497. doi: 10.1016/j.fsigen.2021.102497. Epub 2021 Mar 15. Forensic Sci Int Genet. 2021. PMID: 33740708 Review.
Cited by
-
MANUDB: database and application to retrieve and visualize mammalian NUMTs.Database (Oxford). 2025 Feb 22;2025:baaf009. doi: 10.1093/database/baaf009. Database (Oxford). 2025. PMID: 39985319 Free PMC article.
-
Characterization of Nuclear Mitochondrial Insertions in Canine Genome Assemblies.Genes (Basel). 2024 Oct 14;15(10):1318. doi: 10.3390/genes15101318. Genes (Basel). 2024. PMID: 39457442 Free PMC article.
-
MitSorter: a standalone tool for accurate discrimination of mtDNA and NuMT ONT reads based on differential methylation.Bioinform Adv. 2025 Jul 10;5(1):vbaf135. doi: 10.1093/bioadv/vbaf135. eCollection 2025. Bioinform Adv. 2025. PMID: 40688360 Free PMC article.
-
A Genome-Wide Analysis of Nuclear Mitochondrial DNA Sequences (NUMTs) in Chrysomelidae Species (Coleoptera).Insects. 2025 Feb 2;16(2):150. doi: 10.3390/insects16020150. Insects. 2025. PMID: 40003780 Free PMC article.
References
-
- Bravi C.M., Parson W., Bandelt H.-J. Numts Revisited. In: Bandelt H.-J., Macaulay V., Richards M., editors. Human Mitochondrial DNA and the Evolution of Homo Sapiens. Springer; Berlin/Heidelberg, Germany: 2006. pp. 31–46. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

