Comparative Computational Analysis of Spike Protein Structural Stability in SARS-CoV-2 Omicron Subvariants
- PMID: 38003257
- PMCID: PMC10671153
- DOI: 10.3390/ijms242216069
Comparative Computational Analysis of Spike Protein Structural Stability in SARS-CoV-2 Omicron Subvariants
Abstract
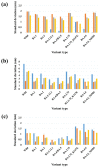
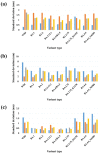
The continuous emergence of new severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants with multiple spike (S) protein mutations pose serious threats to current coronavirus disease 2019 (COVID-19) therapies. A comprehensive understanding of the structural stability of SARS-CoV-2 variants is vital for the development of effective therapeutic strategies as it can offer valuable insights into their potential impact on viral infectivity. S protein mediates a virus' attachment to host cells by binding to angiotensin-converting enzyme 2 (ACE2) through its receptor-binding domain (RBD), and mutations in this protein can affect its stability and binding affinity. We analyzed S protein structural stability in various Omicron subvariants computationally. Notably, the S protein sequences analyzed in this work were obtained directly from our own sample collection. We evaluated the binding free energy between S protein and ACE2 in several complex forms. Additionally, we measured distances between the RBD of each chain in S protein to analyze conformational changes. Unlike most of the prior studies, we analyzed full-length S protein-ACE2 complexes instead of only RBD-ACE2 complexes. Omicron subvariants including BA.1, BA.2, BA.2.12.1, BA.4/BA.5, BA.2.75, BA.2.75_K147E, BA.4.6 and BA.4.6_N658S showed enhanced stability compared to wild type, potentially due to distinct S protein mutations. Among them, BA.2.75 and BA.4.6_N658S exhibited the highest and lowest level of stability, respectively.
Keywords: MD simulation; MM/PBSA; Omicron; SARS-CoV-2.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

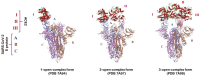

Similar articles
-
Comparative Analysis of Conformational Dynamics and Systematic Characterization of Cryptic Pockets in the SARS-CoV-2 Omicron BA.2, BA.2.75 and XBB.1 Spike Complexes with the ACE2 Host Receptor: Confluence of Binding and Structural Plasticity in Mediating Networks of Conserved Allosteric Sites.Viruses. 2023 Oct 10;15(10):2073. doi: 10.3390/v15102073. Viruses. 2023. PMID: 37896850 Free PMC article.
-
Electrostatic Interactions Are the Primary Determinant of the Binding Affinity of SARS-CoV-2 Spike RBD to ACE2: A Computational Case Study of Omicron Variants.Int J Mol Sci. 2022 Nov 26;23(23):14796. doi: 10.3390/ijms232314796. Int J Mol Sci. 2022. PMID: 36499120 Free PMC article.
-
Temperature-dependent Spike-ACE2 interaction of Omicron subvariants is associated with viral transmission.mBio. 2024 Aug 14;15(8):e0090724. doi: 10.1128/mbio.00907-24. Epub 2024 Jul 2. mBio. 2024. PMID: 38953636 Free PMC article.
-
Molecular and structural insights into SARS-CoV-2 evolution: from BA.2 to XBB subvariants.mBio. 2024 Oct 16;15(10):e0322023. doi: 10.1128/mbio.03220-23. Epub 2024 Sep 16. mBio. 2024. PMID: 39283095 Free PMC article. Review.
-
Interactions of angiotensin-converting enzyme-2 (ACE2) and SARS-CoV-2 spike receptor-binding domain (RBD): a structural perspective.Mol Biol Rep. 2023 Mar;50(3):2713-2721. doi: 10.1007/s11033-022-08193-4. Epub 2022 Dec 23. Mol Biol Rep. 2023. PMID: 36562937 Free PMC article. Review.
Cited by
-
Genetic diversity and genomic epidemiology of SARS-CoV-2 during the first 3 years of the pandemic in Morocco: comprehensive sequence analysis, including the unique lineage B.1.528 in Morocco.Access Microbiol. 2024 Oct 7;6(10):000853.v4. doi: 10.1099/acmi.0.000853.v4. eCollection 2024. Access Microbiol. 2024. PMID: 39376591 Free PMC article.
References
-
- Ou J., Zhou Z., Dai R., Zhang J., Zhao S., Wu X., Lan W., Ren Y., Cui L., Lan Q. V367F mutation in SARS-CoV-2 spike RBD emerging during the early transmission phase enhances viral infectivity through increased human ACE2 receptor binding affinity. J. Virol. 2021;95:e00617-21. doi: 10.1128/JVI.00617-21. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

