Multi-Omics Analysis of Circulating Exosomes in Adherent Long-Term Treated OSA Patients
- PMID: 38003263
- PMCID: PMC10671639
- DOI: 10.3390/ijms242216074
Multi-Omics Analysis of Circulating Exosomes in Adherent Long-Term Treated OSA Patients
Abstract
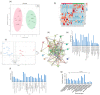
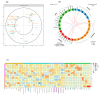
Obstructive sleep apnea (OSA) is a highly prevalent chronic disease affecting nearly a billion people globally and increasing the risk of multi-organ morbidity and overall mortality. However, the mechanisms underlying such adverse outcomes remain incompletely delineated. Extracellular vesicles (exosomes) are secreted by most cells, are involved in both proximal and long-distance intercellular communication, and contribute toward homeostasis under physiological conditions. A multi-omics integrative assessment of plasma-derived exosomes from adult OSA patients prior to and after 1-year adherent CPAP treatment is lacking. We conducted multi-omic integrative assessments of plasma-derived exosomes from adult OSA patients prior to and following 1-year adherent CPAP treatment to identify potential specific disease candidates. Fasting morning plasma exosomes isolated from 12 adult patients with polysomnographically-diagnosed OSA were analyzed before and after 12 months of adherent CPAP therapy (mean ≥ 6 h/night) (OSAT). Exosomes were characterized by flow cytometry, transmission electron microscopy, and nanoparticle tracking analysis. Endothelial cell barrier integrity, wound healing, and tube formation were also performed. Multi-omics analysis for exosome cargos was integrated. Exosomes derived from OSAT improved endothelial permeability and dysfunction as well as significant improvement in tube formation compared with OSA. Multi-omic approaches for OSA circulating exosomes included lipidomic, proteomic, and small RNA (miRNAs) assessments. We found 30 differentially expressed proteins (DEPs), 72 lipids (DELs), and 13 miRNAs (DEMs). We found that the cholesterol metabolism (has04979) pathway is associated with lipid classes in OSA patients. Among the 12 subjects of OSA and OSAT, seven subjects had complete comprehensive exosome cargo information including lipids, proteins, and miRNAs. Multi-omic approaches identify potential signature biomarkers in plasma exosomes that are responsive to adherent OSA treatment. These differentially expressed molecules may also play a mechanistic role in OSA-induced morbidities and their reversibility. Our data suggest that a multi-omic integrative approach might be useful in understanding how exosomes function, their origin, and their potential clinical relevance, all of which merit future exploration in the context of relevant phenotypic variance. Developing an integrated molecular classification should lead to improved diagnostic classification, risk stratification, and patient management of OSA by assigning molecular disease-specific therapies.
Keywords: OSA; exosomes; extracellular vesicles; lipids; miRNAs; multi-omics; omics; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Circulating exosomes in obstructive sleep apnea as phenotypic biomarkers and mechanistic messengers of end-organ morbidity.Respir Physiol Neurobiol. 2018 Oct;256:143-156. doi: 10.1016/j.resp.2017.06.004. Epub 2017 Jul 1. Respir Physiol Neurobiol. 2018. PMID: 28676332 Free PMC article. Review.
-
Circulating Plasma Extracellular Microvesicle MicroRNA Cargo and Endothelial Dysfunction in Children with Obstructive Sleep Apnea.Am J Respir Crit Care Med. 2016 Nov 1;194(9):1116-1126. doi: 10.1164/rccm.201602-0323OC. Am J Respir Crit Care Med. 2016. PMID: 27163713 Free PMC article.
-
Exosome and Macrophage Crosstalk in Sleep-Disordered Breathing-Induced Metabolic Dysfunction.Int J Mol Sci. 2018 Oct 29;19(11):3383. doi: 10.3390/ijms19113383. Int J Mol Sci. 2018. PMID: 30380647 Free PMC article. Review.
-
Plasma exosomes in OSA patients promote endothelial senescence: effect of long-term adherent continuous positive airway pressure.Sleep. 2020 Feb 13;43(2):zsz217. doi: 10.1093/sleep/zsz217. Sleep. 2020. PMID: 31552414 Free PMC article.
-
Circulating exosomes potentiate tumor malignant properties in a mouse model of chronic sleep fragmentation.Oncotarget. 2016 Aug 23;7(34):54676-54690. doi: 10.18632/oncotarget.10578. Oncotarget. 2016. PMID: 27419627 Free PMC article.
Cited by
-
Childhood Asthma Biomarkers Derived from Plasma and Saliva Exosomal miRNAs.Int J Mol Sci. 2025 Jul 22;26(15):7043. doi: 10.3390/ijms26157043. Int J Mol Sci. 2025. PMID: 40806181 Free PMC article.
-
Algorithms and tools for data-driven omics integration to achieve multilayer biological insights: a narrative review.J Transl Med. 2025 Apr 10;23(1):425. doi: 10.1186/s12967-025-06446-x. J Transl Med. 2025. PMID: 40211300 Free PMC article. Review.
-
Recent Progress in Omics Studies of Sleep and Circadian Phenotypes.Curr Sleep Med Rep. 2025 Dec;11(1):17. doi: 10.1007/s40675-025-00335-x. Epub 2025 Apr 15. Curr Sleep Med Rep. 2025. PMID: 40321983
-
Revolutionizing Sleep Health: The Emergence and Impact of Personalized Sleep Medicine.J Pers Med. 2024 Jun 4;14(6):598. doi: 10.3390/jpm14060598. J Pers Med. 2024. PMID: 38929819 Free PMC article. Review.
References
-
- Benjafield A.V., Ayas N.T., Eastwood P.R., Heinzer R., Ip M.S.M., Morrell M.J., Nunez C.M., Patel S.R., Penzel T., Pepin J.L., et al. Estimation of the global prevalence and burden of obstructive sleep apnoea: A literature-based analysis. Lancet Respir. Med. 2019;7:687–698. doi: 10.1016/S2213-2600(19)30198-5. - DOI - PMC - PubMed
-
- Ding F., Cotton-Clay A., Fava L., Easwar V., Kinsolving A., Kahn P., Rama A., Kushida C. Polysomnographic validation of an under-mattress monitoring device in estimating sleep architecture and obstructive sleep apnea in adults. Sleep Med. 2022;96:20–27. doi: 10.1016/j.sleep.2022.04.010. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

