Linear Dichroism Measurements for the Study of Protein-DNA Interactions
- PMID: 38003280
- PMCID: PMC10671323
- DOI: 10.3390/ijms242216092
Linear Dichroism Measurements for the Study of Protein-DNA Interactions
Abstract
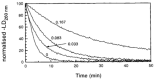
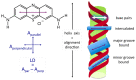
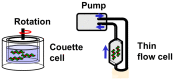
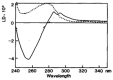
Linear dichroism (LD) is a differential polarized light absorption spectroscopy used for studying filamentous molecules such as DNA and protein filaments. In this study, we review the applications of LD for the analysis of DNA-protein interactions. LD signals can be measured in a solution by aligning the sample using flow-induced shear force or a strong electric field. The signal generated is related to the local orientation of chromophores, such as DNA bases, relative to the filament axis. LD can thus assess the tilt and roll of DNA bases and distinguish intercalating from groove-binding ligands. The intensity of the LD signal depends upon the degree of macroscopic orientation. Therefore, DNA shortening and bending can be detected by a decrease in LD signal intensity. As examples of LD applications, we present a kinetic study of DNA digestion by restriction enzymes and structural analyses of homologous recombination intermediates, i.e., RecA and Rad51 recombinase complexes with single-stranded DNA. LD shows that the DNA bases in these complexes are preferentially oriented perpendicular to the filament axis only in the presence of activators, suggesting the importance of organized base orientation for the reaction. LD measurements detect DNA bending by the CRP transcription activator protein, as well as by the UvrB DNA repair protein. LD can thus provide information about the structures of protein-DNA complexes under various conditions and in real time.
Keywords: DNA/protein complex; Rad51; RecA; UvrB nucleotide excision repair protein; catabolism activator protein (CAP); cyclic AMP receptor protein (CRP); homologous recombination; linear dichroism (LD); restriction enzyme; transcription regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Linear dichroism reveals the perpendicular orientation of DNA bases in the RecA and Rad51 recombinase filaments: A possible mechanism for the strand exchange reaction.Chirality. 2024 Apr;36(4):e23664. doi: 10.1002/chir.23664. Chirality. 2024. PMID: 38561319
-
Coordination and internal exchange of two DNA molecules in a RecA filament in the presence of hydrolysing ATP. Information on ATP-RecA-DNA structure from linear dichroism spectroscopy.Eur J Biochem. 1992 Nov 15;210(1):87-92. doi: 10.1111/j.1432-1033.1992.tb17394.x. Eur J Biochem. 1992. PMID: 1446687
-
Linear dichroism study of RecA-DNA complexes. Structural evidence and binding stoichiometries.J Biol Chem. 1987 Jun 15;262(17):8109-11. J Biol Chem. 1987. PMID: 3597365
-
Analysing DNA complexes by circular and linear dichroism.J Mol Recognit. 1994 Jun;7(2):141-55. doi: 10.1002/jmr.300070211. J Mol Recognit. 1994. PMID: 7826674 Review.
-
Homologous genetic recombination as an intrinsic dynamic property of a DNA structure induced by RecA/Rad51-family proteins: a possible advantage of DNA over RNA as genomic material.Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8425-32. doi: 10.1073/pnas.111005198. Proc Natl Acad Sci U S A. 2001. PMID: 11459985 Free PMC article. Review.
Cited by
-
Human Rad51 Protein Requires Higher Concentrations of Calcium Ions for D-Loop Formation than for Oligonucleotide Strand Exchange.Int J Mol Sci. 2024 Mar 24;25(7):3633. doi: 10.3390/ijms25073633. Int J Mol Sci. 2024. PMID: 38612444 Free PMC article.
References
-
- Norden B., Elvingson C., Kubista M., Sjöberg B., Ryberg H., Ryberg M., Mortensen K., Takahashi M. Structure of RecA-DNA complexes studied by combination of linear dichroism and small angle neutron scattering measurements on flow-oriented samples. J. Mol. Biol. 1992;226:1175–1191. doi: 10.1016/0022-2836(92)91060-3. - DOI - PubMed
-
- Tuite E., Sehlstedt U., Hagmar P., Norden B., Takahashi M. Effects of minor and major groove binding drugs and intercalators on the DNA association of minor-groove binding proteins RecA and DNaseI detected by flow linear dichroism. Eur. J. Biochem. 1997;243:482–492. doi: 10.1111/j.1432-1033.1997.0482a.x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

