Identification of a Pentasaccharide Lead Compound with High Affinity to the SARS-CoV-2 Spike Protein via In Silico Screening
- PMID: 38003304
- PMCID: PMC10671481
- DOI: 10.3390/ijms242216115
Identification of a Pentasaccharide Lead Compound with High Affinity to the SARS-CoV-2 Spike Protein via In Silico Screening
Abstract
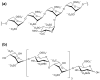
The spike (S) protein on the surface of the SARS-CoV-2 virus is critical to mediate fusion with the host cell membrane through interaction with angiotensin-converting enzyme 2 (ACE2). Additionally, heparan sulfate (HS) on the host cell surface acts as an attachment factor to facilitate the binding of the S receptor binding domain (RBD) to the ACE2 receptor. Aiming at interfering with the HS-RBD interaction to protect against SARS-CoV-2 infection, we have established a pentasaccharide library composed of 14,112 compounds covering the possible sulfate substitutions on the three sugar units (GlcA, IdoA, and GlcN) of HS. The library was used for virtual screening against RBD domains of SARS-CoV-2. Molecular modeling was carried out to evaluate the potential antiviral properties of the top-hit pentasaccharide focusing on the interactive regions around the interface of RBD-HS-ACE2. The lead pentasaccharide with the highest affinity for RBD was analyzed via drug-likeness calculations, showing better predicted druggable profiles than those currently reported for RBD-binding HS mimetics. The results provide significant information for the development of HS-mimetics as anti-SARS-CoV-2 agents.
Keywords: RBD; SARS-CoV-2; heparan sulfate; protein binding; virtual screening.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

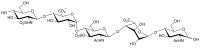






Similar articles
-
AI Promoted Virtual Screening, Structure-Based Hit Optimization, and Synthesis of Novel COVID-19 S-RBD Domain Inhibitors.J Chem Inf Model. 2024 Nov 25;64(22):8562-8585. doi: 10.1021/acs.jcim.4c01110. Epub 2024 Nov 13. J Chem Inf Model. 2024. PMID: 39535926 Free PMC article.
-
The accomplices: Heparan sulfates and N-glycans foster SARS-CoV-2 spike:ACE2 receptor binding and virus priming.Proc Natl Acad Sci U S A. 2024 Oct 22;121(43):e2404892121. doi: 10.1073/pnas.2404892121. Epub 2024 Oct 14. Proc Natl Acad Sci U S A. 2024. PMID: 39401361 Free PMC article.
-
Identification of SARS-CoV-2 Receptor Binding Inhibitors by In Vitro Screening of Drug Libraries.Molecules. 2021 May 27;26(11):3213. doi: 10.3390/molecules26113213. Molecules. 2021. PMID: 34072087 Free PMC article.
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
-
Interactions of angiotensin-converting enzyme-2 (ACE2) and SARS-CoV-2 spike receptor-binding domain (RBD): a structural perspective.Mol Biol Rep. 2023 Mar;50(3):2713-2721. doi: 10.1007/s11033-022-08193-4. Epub 2022 Dec 23. Mol Biol Rep. 2023. PMID: 36562937 Free PMC article. Review.
Cited by
-
Special Issue: "Rational Design and Synthesis of Bioactive Molecules".Int J Mol Sci. 2024 Sep 14;25(18):9927. doi: 10.3390/ijms25189927. Int J Mol Sci. 2024. PMID: 39337415 Free PMC article.
-
Identification of an Unnatural Sulfated Monosaccharide as a High-Affinity Ligand for Pan-Variant Targeting of SARS-CoV-2 Spike Glycoprotein.ACS Chem Biol. 2025 Jun 20;20(6):1394-1405. doi: 10.1021/acschembio.5c00206. Epub 2025 May 13. ACS Chem Biol. 2025. PMID: 40358361 Free PMC article.
References
-
- Ghai R.R., Carpenter A., Liew A.Y., Martin K.B., Herring M.K., Gerber S.I., Hall A.J., Sleeman J.M., VonDobschuetz S., Behravesh C.B. Animal Reservoirs and Hosts for Emerging Alphacoronaviruses and Betacoronaviruses. Emerg. Infect. Dis. 2021;27:1015–1022. doi: 10.3201/eid2704.203945. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 2021YFC2103100/the National Key R&D Program of China
- 32201053/the National Natural Science Foundation of China
- 21530009117/Beijing Advanced Innovation Center for Soft Matter Science and Engineering
- 3100012222222/Beijing Institute of Technology Research Fund Program for Young Scholars
- 2021-01094/Swedish Research Council
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

