CRISPR/Cas9 Mutagenesis through Introducing a Nanoparticle Complex Made of a Cationic Polymer and Nucleic Acids into Maize Protoplasts
- PMID: 38003326
- PMCID: PMC10671792
- DOI: 10.3390/ijms242216137
CRISPR/Cas9 Mutagenesis through Introducing a Nanoparticle Complex Made of a Cationic Polymer and Nucleic Acids into Maize Protoplasts
Abstract
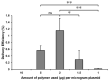
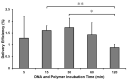
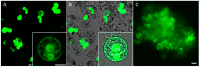
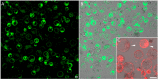
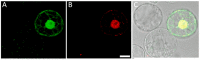
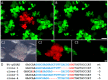
Presently, targeted gene mutagenesis attracts increasing attention both in plant research and crop improvement. In these approaches, successes are largely dependent on the efficiency of the delivery of gene editing components into plant cells. Here, we report the optimization of the cationic polymer poly(2-hydroxypropylene imine) (PHPI)-mediated delivery of plasmid DNAs, or single-stranded oligonucleotides labelled with Cyanine3 (Cy3) or 6-Carboxyfluorescein (6-FAM)-fluorescent dyes into maize protoplasts. Co-delivery of the GFP-expressing plasmid and the Cy3-conjugated oligonucleotides has resulted in the cytoplasmic and nuclear accumulation of the green fluorescent protein and a preferential nuclear localization of oligonucleotides. We show the application of nanoparticle complexes, i.e., "polyplexes" that comprise cationic polymers and nucleic acids, for CRISPR/Cas9 editing of maize cells. Knocking out the functional EGFP gene in transgenic maize protoplasts was achieved through the co-delivery of plasmids encoding components of the editing factors Cas9 (pFGC-pcoCas9) and gRNA (pZmU3-gRNA) after complexing with a cationic polymer (PHPI). Several edited microcalli were identified based on the lack of a GFP fluorescence signal. Multi-base and single-base deletions in the EGFP gene were confirmed using Sanger sequencing. The presented results support the use of the PHPI cationic polymer in plant protoplast-mediated genome editing approaches.
Keywords: CRISPR/Cas9; DNA oligonucleotide; DsRed fluorescent marker protein; GFP expression; maize protoplasts; mutagenesis; nanoparticle; plasmid uptake; poly (2-hydroxypropylene imine); polyplexes.
Conflict of interest statement
The authors declare no conflict of interest. Author I. Nagy was employed by the company SeqOmics Biotechnology Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Jiang Y., Sun K., An X. CRISPR/Cas System: Applications and Prospects for Maize Improvement. ACS Agric. Sci. Technol. 2022;2:174–183. doi: 10.1021/acsagscitech.1c00253. - DOI
-
- Gordon-Kamm W., Barone P., Svitashev S., Sander J.D., Kumar S., Jones T. Strategies for CRISPR/Cas9-Mediated Genome Editing: From Delivery to Production of Modified Plants. Burleigh Dodds Science Publishing; Sawston, UK: 2021.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

