Hsp70 and Hsp90 Elaborately Regulate RNAi Efficiency in Plutella xylostella
- PMID: 38003357
- PMCID: PMC10671170
- DOI: 10.3390/ijms242216167
Hsp70 and Hsp90 Elaborately Regulate RNAi Efficiency in Plutella xylostella
Abstract
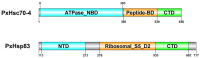
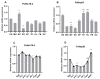
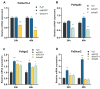
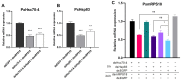
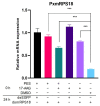
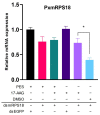
Heat-shock proteins (HSPs) serve as molecular chaperones in the RNA interference (RNAi) pathway of eukaryotic organisms. In model organisms, Hsp70 and Hsp90 facilitate the folding and remodeling of the client protein Argonaute (Ago). However, the specific function of HSPs in the RNAi pathway of Plutella xylostella (L.) (Lepidoptera: Plutellidae) remains unknown. In this study, we identified and analyzed the coding sequences of PxHsc70-4 and PxHsp83 (also known as PxHsp90). Both PxHsc70-4 and PxHsp83 exhibited three conserved domains that covered a massive portion of their respective regions. The knockdown or inhibition of PxHsc70-4 and PxHsp83 in vitro resulted in a significant increase in the gene expression of the dsRNA-silenced reporter gene PxmRPS18, leading to a decrease in its RNAi efficiency. Interestingly, the overexpression of PxHsc70-4 and PxHsp83 in DBM, Sf9, and S2 cells resulted in an increase in the bioluminescent activity of dsRNA-silenced luciferase, indicating a decrease in its RNAi efficiency via the overexpression of Hsp70/Hsp90. Furthermore, the inhibition of PxHsc70-4 and PxHsp83 in vivo resulted in a significant increase in the gene expression of PxmRPS18. These findings demonstrated the essential involvement of a specific quantity of Hsc70-4 and Hsp83 in the siRNA pathway in P. xylostella. Our study offers novel insights into the roles played by HSPs in the siRNA pathway in lepidopteran insects.
Keywords: Hsp70; Hsp90; Plutella xylostella; RNA interference.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Molecular Characterization and the Function of Argonaute3 in RNAi Pathway of Plutella xylostella.Int J Mol Sci. 2018 Apr 20;19(4):1249. doi: 10.3390/ijms19041249. Int J Mol Sci. 2018. PMID: 29677157 Free PMC article.
-
RNAi-dependent heterochromatin assembly in fission yeast Schizosaccharomyces pombe requires heat-shock molecular chaperones Hsp90 and Mas5.Epigenetics Chromatin. 2018 Jun 4;11(1):26. doi: 10.1186/s13072-018-0199-8. Epigenetics Chromatin. 2018. PMID: 29866182 Free PMC article.
-
A direct role for Hsp90 in pre-RISC formation in Drosophila.Nat Struct Mol Biol. 2010 Aug;17(8):1024-6. doi: 10.1038/nsmb.1875. Epub 2010 Jul 18. Nat Struct Mol Biol. 2010. PMID: 20639883
-
Role of Argonaute proteins in RNAi pathway in Plutella xylostella: A review.Gene. 2024 Apr 20;903:148195. doi: 10.1016/j.gene.2024.148195. Epub 2024 Jan 29. Gene. 2024. PMID: 38295911 Review.
-
Hsp90 and Hsp70 chaperones: Collaborators in protein remodeling.J Biol Chem. 2019 Feb 8;294(6):2109-2120. doi: 10.1074/jbc.REV118.002806. Epub 2018 Nov 6. J Biol Chem. 2019. PMID: 30401745 Free PMC article. Review.
Cited by
-
Environmental factors affecting RNAi efficacy: Temperature but not plant cultivar influences Colorado potato beetle's response to insecticidal dsRNA.Insect Mol Biol. 2025 Aug;34(4):581-592. doi: 10.1111/imb.12996. Epub 2025 May 23. Insect Mol Biol. 2025. PMID: 40410128 Free PMC article.
-
Recent advances in understanding of the mechanisms of RNA interference in insects.Insect Mol Biol. 2025 Aug;34(4):491-504. doi: 10.1111/imb.12941. Epub 2024 Jul 3. Insect Mol Biol. 2025. PMID: 38957135 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
- 2017YFD0200400/National Key R&D Program of China
- 2018NZ01010013/Special Key Project of Fujian Province
- KRA16001A/fund of "111" program
- KJG18018A/fund of Joint International Laboratory, China
- KXB16014A/Fujian Agriculture and Forestry University International Science and Tech- 424 nology Cooperation and Exchange Program
LinkOut - more resources
Full Text Sources
Miscellaneous

