Nanoscale Bilirubin Analysis in Translational Research and Precision Medicine by the Recombinant Protein HUG
- PMID: 38003479
- PMCID: PMC10671013
- DOI: 10.3390/ijms242216289
Nanoscale Bilirubin Analysis in Translational Research and Precision Medicine by the Recombinant Protein HUG
Abstract
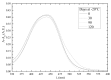
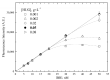
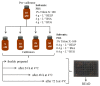
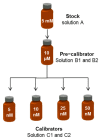
Bilirubin is a toxicological biomarker for hemolysis and liver diseases. The current automated diazo method used in clinical chemistry has limited applicability in rodent models and cannot be used in small animals relevant to toxicology, microphysiological systems, cell cultures, and kinetic studies. Here, we present a versatile fluorometric method for nanoscale analysis of bilirubin based on its highly specific binding to the recombinant bifunctional protein HELP-UnaG (HUG). The assay is sensitive (LoQ = 1.1 nM), accurate (4.5% relative standard error), and remarkably robust, allowing analysis at pH 7.4-9.5, T = 25-37 °C, in various buffers, and in the presence of 0.4-4 mg × L-1 serum albumin or 30% DMSO. It allows repeated measurements of bilirubinemia in murine models and small animals, fostering the 3Rs principle. The assay determines bilirubin in human plasma with a relative standard error of 6.7% at values that correlate and agree with the standard diazo method. Furthermore, it detects differences in human bilirubinemia related to sex and UGT1A1 polymorphisms, thus demonstrating its suitability for the uniform assessment of bilirubin at the nanoscale in translational and precision medicine.
Keywords: bilirubin; biomarker; biomedical diagnostics; calibration; fluorescent method; high-throughput assay.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








Similar articles
-
Standardized lab-scale production of the recombinant fusion protein HUG for the nanoscale analysis of bilirubin.MethodsX. 2024 Oct 17;13:103001. doi: 10.1016/j.mex.2024.103001. eCollection 2024 Dec. MethodsX. 2024. PMID: 39469066 Free PMC article.
-
The HELP-UnaG Fusion Protein as a Bilirubin Biosensor: From Theory to Mature Technological Development.Molecules. 2025 Jan 21;30(3):439. doi: 10.3390/molecules30030439. Molecules. 2025. PMID: 39942546 Free PMC article. Review.
-
Macromolecular and Solution Properties of the Recombinant Fusion Protein HUG.Biomacromolecules. 2022 Aug 8;23(8):3336-3348. doi: 10.1021/acs.biomac.2c00447. Epub 2022 Jul 25. Biomacromolecules. 2022. PMID: 35876275 Free PMC article.
-
Bilirubin conjugation, reflected by conjugated bilirubin fractions, in glucose-6-phosphate dehydrogenase-deficient neonates: a determining factor in the pathogenesis of hyperbilirubinemia.Pediatrics. 1998 Sep;102(3):E37. doi: 10.1542/peds.102.3.e37. Pediatrics. 1998. PMID: 9724685 Clinical Trial.
-
The Importance of Hemolysis and Its Clinical Detection in Neonates with Hyperbilirubinemia.Curr Pediatr Rev. 2017;13(3):193-198. doi: 10.2174/1573396313666170807121444. Curr Pediatr Rev. 2017. PMID: 28782473 Review.
Cited by
-
1,4-dihydroxy quininib activates ferroptosis pathways in metastatic uveal melanoma and reveals a novel prognostic biomarker signature.Cell Death Discov. 2024 Feb 10;10(1):70. doi: 10.1038/s41420-023-01773-8. Cell Death Discov. 2024. PMID: 38341410 Free PMC article.
-
Standardized lab-scale production of the recombinant fusion protein HUG for the nanoscale analysis of bilirubin.MethodsX. 2024 Oct 17;13:103001. doi: 10.1016/j.mex.2024.103001. eCollection 2024 Dec. MethodsX. 2024. PMID: 39469066 Free PMC article.
-
Preparation and validation of nanomolar aqueous bilirubin standard solutions.MethodsX. 2024 Dec 20;14:103123. doi: 10.1016/j.mex.2024.103123. eCollection 2025 Jun. MethodsX. 2024. PMID: 39830880 Free PMC article.
-
The HELP-UnaG Fusion Protein as a Bilirubin Biosensor: From Theory to Mature Technological Development.Molecules. 2025 Jan 21;30(3):439. doi: 10.3390/molecules30030439. Molecules. 2025. PMID: 39942546 Free PMC article. Review.
-
Combined fluorometric analysis of biliverdin and bilirubin by the recombinant protein HUG.MethodsX. 2024 Sep 25;13:102979. doi: 10.1016/j.mex.2024.102979. eCollection 2024 Dec. MethodsX. 2024. PMID: 39430781 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

