Structural Insights into Protein-Aptamer Recognitions Emerged from Experimental and Computational Studies
- PMID: 38003510
- PMCID: PMC10671752
- DOI: 10.3390/ijms242216318
Structural Insights into Protein-Aptamer Recognitions Emerged from Experimental and Computational Studies
Abstract
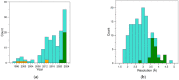
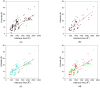
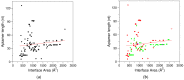
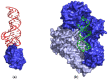
Aptamers are synthetic nucleic acids that are developed to target with high affinity and specificity chemical entities ranging from single ions to macromolecules and present a wide range of chemical and physical properties. Their ability to selectively bind proteins has made these compounds very attractive and versatile tools, in both basic and applied sciences, to such an extent that they are considered an appealing alternative to antibodies. Here, by exhaustively surveying the content of the Protein Data Bank (PDB), we review the structural aspects of the protein-aptamer recognition process. As a result of three decades of structural studies, we identified 144 PDB entries containing atomic-level information on protein-aptamer complexes. Interestingly, we found a remarkable increase in the number of determined structures in the last two years as a consequence of the effective application of the cryo-electron microscopy technique to these systems. In the present paper, particular attention is devoted to the articulated architectures that protein-aptamer complexes may exhibit. Moreover, the molecular mechanism of the binding process was analyzed by collecting all available information on the structural transitions that aptamers undergo, from their protein-unbound to the protein-bound state. The contribution of computational approaches in this area is also highlighted.
Keywords: NMR; X-ray crystallography; allostery; aptamer; cryo-electron microscopy; crystal structure; molecular dynamics; protein data bank; protein–aptamer interface; ternary complex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Characterization of aptamer-protein complexes by X-ray crystallography and alternative approaches.Int J Mol Sci. 2012;13(8):10537-10552. doi: 10.3390/ijms130810537. Epub 2012 Aug 22. Int J Mol Sci. 2012. PMID: 22949878 Free PMC article. Review.
-
Impact of the Position of the Chemically Modified 5-Furyl-2'-Deoxyuridine Nucleoside on the Thrombin DNA Aptamer-Protein Complex: Structural Insights into Aptamer Response from MD Simulations.Molecules. 2019 Aug 10;24(16):2908. doi: 10.3390/molecules24162908. Molecules. 2019. PMID: 31405145 Free PMC article.
-
Investigations on the interface of nucleic acid aptamers and binding targets.Analyst. 2018 Nov 5;143(22):5317-5338. doi: 10.1039/c8an01467a. Analyst. 2018. PMID: 30357118 Review.
-
Energy landscape of aptamer/protein complexes studied by single-molecule force spectroscopy.Chem Asian J. 2007 Feb 5;2(2):284-9. doi: 10.1002/asia.200600230. Chem Asian J. 2007. PMID: 17441163
-
Investigating the malleability of RNA aptamers.Methods. 2013 Sep 15;63(2):178-87. doi: 10.1016/j.ymeth.2013.03.016. Epub 2013 Mar 25. Methods. 2013. PMID: 23535583
Cited by
-
Targeting eIF4A with RNA Aptamers Enhances Salt Stress Tolerance in Rice Through Modulation of Translation Initiation.Rice (N Y). 2025 Jul 7;18(1):62. doi: 10.1186/s12284-025-00819-y. Rice (N Y). 2025. PMID: 40622653 Free PMC article.
-
Computational Frontiers in Aptamer-Based Nanomedicine for Precision Therapeutics: A Comprehensive Review.ACS Omega. 2024 Jun 10;9(25):26838-26862. doi: 10.1021/acsomega.4c02466. eCollection 2024 Jun 25. ACS Omega. 2024. PMID: 38947800 Free PMC article. Review.
-
Recent Advances in Aptamer-Based Biosensors for Bacterial Detection.Biosensors (Basel). 2024 Apr 23;14(5):210. doi: 10.3390/bios14050210. Biosensors (Basel). 2024. PMID: 38785684 Free PMC article. Review.
-
Aptamer based immunotherapy: a potential solid tumor therapeutic.Front Immunol. 2025 Feb 17;16:1536569. doi: 10.3389/fimmu.2025.1536569. eCollection 2025. Front Immunol. 2025. PMID: 40034705 Free PMC article. Review.
-
Aptamers: Functional-Structural Studies and Biomedical Applications 2.0.Int J Mol Sci. 2024 Apr 12;25(8):4279. doi: 10.3390/ijms25084279. Int J Mol Sci. 2024. PMID: 38673864 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

