Characterization of the Moroccan Barley Germplasm Preserved in the Polish Genebank as a First Step towards Selecting Forms with Increased Drought Tolerance
- PMID: 38003539
- PMCID: PMC10671370
- DOI: 10.3390/ijms242216350
Characterization of the Moroccan Barley Germplasm Preserved in the Polish Genebank as a First Step towards Selecting Forms with Increased Drought Tolerance
Abstract
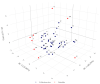
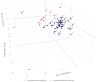
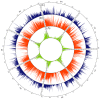
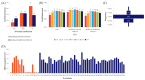
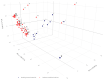
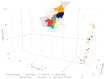
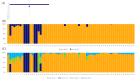
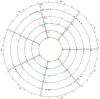
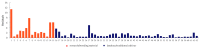
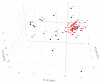
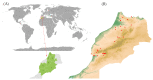
In marginal, arid, and semi-arid areas of Morocco, crops are often exposed to multiple abiotic and biotic stresses that have a major impact on yield. Farmer-maintained Moroccan landraces have been shaped by the impact of very strong selection pressures, gradually adapting to the local ecosystem and obsolete low-input agricultural practices without improvement towards high yield and quality. Considering the increasing threat of drought in Poland, it is necessary to introduce germplasm with tolerance to water deficit into barley breeding programs. The aim of this research was a DArTseq-based genetic characterization of a collection of germplasm of Moroccan origin, conserved in the Polish genebank. The results showed that all conserved landraces have a high level of heterogeneity and their gene pool is different from the material developed by Polish breeders. Based on the analysis of eco-geographical data, locations with extremely different intensities of drought stress were selected. A total of 129 SNPs unique to accessions from these locations were identified. In the neighborhood of the clusters of unique SNPs on chromosomes 5H and 6H, genes that may be associated with plant response to drought stress were identified. The results obtained may provide a roadmap for further research to support Polish barley breeding for increased drought tolerance.
Keywords: DArTseq; SNP; barley; diversity; drought tolerance; germplasm.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

Similar articles
-
Drought Stress Tolerance in Wheat and Barley: Advances in Physiology, Breeding and Genetics Research.Int J Mol Sci. 2019 Jun 27;20(13):3137. doi: 10.3390/ijms20133137. Int J Mol Sci. 2019. PMID: 31252573 Free PMC article. Review.
-
Natural Variation Uncovers Candidate Genes for Barley Spikelet Number and Grain Yield under Drought Stress.Genes (Basel). 2020 May 11;11(5):533. doi: 10.3390/genes11050533. Genes (Basel). 2020. PMID: 32403266 Free PMC article.
-
Meta-analysis of major QTL for abiotic stress tolerance in barley and implications for barley breeding.Planta. 2017 Feb;245(2):283-295. doi: 10.1007/s00425-016-2605-4. Epub 2016 Oct 11. Planta. 2017. PMID: 27730410
-
Genetic basis of drought tolerance during seed germination in barley.PLoS One. 2018 Nov 2;13(11):e0206682. doi: 10.1371/journal.pone.0206682. eCollection 2018. PLoS One. 2018. PMID: 30388157 Free PMC article.
-
Drought and salt tolerances in wild relatives for wheat and barley improvement.Plant Cell Environ. 2010 Apr;33(4):670-85. doi: 10.1111/j.1365-3040.2009.02107.x. Epub 2010 Feb 5. Plant Cell Environ. 2010. PMID: 20040064 Review.
References
-
- Sloan S., Jenkins C.N., Joppa L.N., Gaveau D.L., Laurance W.F. Remaining natural vegetation in the global biodiversity hotspots. Biol. Conserv. 2014;177:12–24. doi: 10.1016/j.biocon.2014.05.027. - DOI
-
- Noss R.F., Platt W.J., Sorrie B.A., Weakley A.S., Means D.B., Costanza J., Peet R.K. How global biodiversity hotspots may go unrecognized: Lessons from the North American Coastal Plain. Divers. Distrib. 2015;21:236–244. doi: 10.1111/ddi.12278. - DOI
-
- Habel J.C., Rasche L., Schneider U.A., Engler J.O., Schmid E., Rödder D., Meyer S.T., Trapp N., Sos del Diego R., Eggermont H. Final countdown for biodiversity hotspots. Conserv. Lett. 2019;12:e12668. doi: 10.1111/conl.12668. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

