Inconspicuous Yet Indispensable: The Coronavirus Spike Transmembrane Domain
- PMID: 38003610
- PMCID: PMC10671605
- DOI: 10.3390/ijms242216421
Inconspicuous Yet Indispensable: The Coronavirus Spike Transmembrane Domain
Abstract
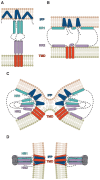
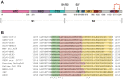
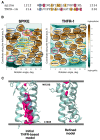
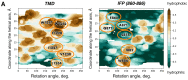
Membrane-spanning portions of proteins' polypeptide chains are commonly known as their transmembrane domains (TMDs). The structural organisation and dynamic behaviour of TMDs from proteins of various families, be that receptors, ion channels, enzymes etc., have been under scrutiny on the part of the scientific community for the last few decades. The reason for such attention is that, apart from their obvious role as an "anchor" in ensuring the correct orientation of the protein's extra-membrane domains (in most cases functionally important), TMDs often actively and directly contribute to the operation of "the protein machine". They are capable of transmitting signals across the membrane, interacting with adjacent TMDs and membrane-proximal domains, as well as with various ligands, etc. Structural data on TMD arrangement are still fragmentary at best due to their complex molecular organisation as, most commonly, dynamic oligomers, as well as due to the challenges related to experimental studies thereof. Inter alia, this is especially true for viral fusion proteins, which have been the focus of numerous studies for quite some time, but have provoked unprecedented interest in view of the SARS-CoV-2 pandemic. However, despite numerous structure-centred studies of the spike (S) protein effectuating target cell entry in coronaviruses, structural data on the TMD as part of the entire spike protein are still incomplete, whereas this segment is known to be crucial to the spike's fusogenic activity. Therefore, in attempting to bring together currently available data on the structure and dynamics of spike proteins' TMDs, the present review aims to tackle a highly pertinent task and contribute to a better understanding of the molecular mechanisms underlying virus-mediated fusion, also offering a rationale for the design of novel efficacious methods for the treatment of infectious diseases caused by SARS-CoV-2 and related viruses.
Keywords: helix–helix interface; membrane protein structure; molecular modelling; protein–protein interactions in membrane; transmembrane helical trimer; viral fusion protein; viral protein acylation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Basso L.G.M., Zeraik A.E., Felizatti A.P., Costa-Filho A.J. Membranotropic and biological activities of the membrane fusion peptides from SARS-CoV spike glycoprotein: The importance of the complete internal fusion peptide domain. Biochim. Biophys. Acta Biomembr. 2021;1863:183697. doi: 10.1016/j.bbamem.2021.183697. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

