Self-Entrapment of Antimicrobial Peptides in Silica Particles for Stable and Effective Antimicrobial Peptide Delivery System
- PMID: 38003614
- PMCID: PMC10671715
- DOI: 10.3390/ijms242216423
Self-Entrapment of Antimicrobial Peptides in Silica Particles for Stable and Effective Antimicrobial Peptide Delivery System
Abstract
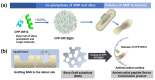
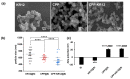
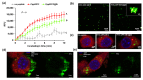
Antimicrobial peptides (AMPs) have emerged as a promising solution to tackle bacterial infections and combat antibiotic resistance. However, their vulnerability to protease degradation and toxicity towards mammalian cells has hindered their clinical application. To overcome these challenges, our study aims to develop a method to enhance the stability and safety of AMPs applicable to effective drug-device combination products. The KR12 antimicrobial peptide was chosen, and in order to further enhance its delivery and efficacy the human immunodeficiency virus TAT protein-derived cell-penetrating peptide (CPP) was fused to form CPP-KR12. A new product, CPP-KR12@Si, was developed by forming silica particles with self-entrapped CPP-KR12 peptide using biomimetic silica precipitability because of its cationic nature. Peptide delivery from CPP-KR12@Si to bacteria and cells was observed at a slightly delivered rate, with improved stability against trypsin treatment and a reduction in cytotoxicity compared to CPP-KR12. Finally, the antimicrobial potential of the CPP-KR12@Si/bone graft substitute (BGS) combination product was demonstrated. CPP-KR12 is coated in the form of submicron-sized particles on the surface of the BGS. Self-entrapped AMP in silica nanoparticles is a safe and effective AMP delivery method that will be useful for developing a drug-device combination product for tissue regeneration.
Keywords: antimicrobial peptide; biomimetic silica deposition; cell penetrating peptide; drug delivery; drug device combination; silica forming peptide.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


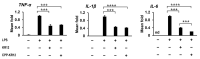


Similar articles
-
Enhancing the Antimicrobial Properties of Peptides through Cell-Penetrating Peptide Conjugation: A Comprehensive Assessment.Int J Mol Sci. 2023 Nov 24;24(23):16723. doi: 10.3390/ijms242316723. Int J Mol Sci. 2023. PMID: 38069046 Free PMC article.
-
Probing the disparate effects of arginine and lysine residues on antimicrobial peptide/bilayer association.Biochim Biophys Acta Biomembr. 2017 Oct;1859(10):1941-1950. doi: 10.1016/j.bbamem.2017.06.002. Epub 2017 Jun 3. Biochim Biophys Acta Biomembr. 2017. PMID: 28583830 Free PMC article.
-
Iron Oxide Nanoparticles with Supramolecular Ureido-Pyrimidinone Coating for Antimicrobial Peptide Delivery.Int J Mol Sci. 2023 Sep 27;24(19):14649. doi: 10.3390/ijms241914649. Int J Mol Sci. 2023. PMID: 37834098 Free PMC article.
-
Antimicrobial peptides with cell-penetrating peptide properties and vice versa.Eur Biophys J. 2011 Apr;40(4):387-97. doi: 10.1007/s00249-011-0682-7. Epub 2011 Feb 19. Eur Biophys J. 2011. PMID: 21336522 Review.
-
Antimicrobial peptides with cell-penetrating activity as prophylactic and treatment drugs.Biosci Rep. 2022 Sep 30;42(9):BSR20221789. doi: 10.1042/BSR20221789. Biosci Rep. 2022. PMID: 36052730 Free PMC article. Review.
Cited by
-
Biominerals and Bioinspired Materials in Biosensing: Recent Advancements and Applications.Int J Mol Sci. 2024 Apr 25;25(9):4678. doi: 10.3390/ijms25094678. Int J Mol Sci. 2024. PMID: 38731897 Free PMC article. Review.
-
Bioinformation and Monitoring Technology for Environmental DNA Analysis: A Review.Biosensors (Basel). 2025 Aug 1;15(8):494. doi: 10.3390/bios15080494. Biosensors (Basel). 2025. PMID: 40862956 Free PMC article. Review.
-
Antimicrobial and Hemostatic Diatom Biosilica Composite Sponge.Antibiotics (Basel). 2024 Jul 30;13(8):714. doi: 10.3390/antibiotics13080714. Antibiotics (Basel). 2024. PMID: 39200014 Free PMC article.
-
Special Issue "Recent Advances in Nanoparticles in Molecular Biology".Int J Mol Sci. 2025 Jun 30;26(13):6321. doi: 10.3390/ijms26136321. Int J Mol Sci. 2025. PMID: 40650098 Free PMC article.
-
Origami of KR-12 Designed Antimicrobial Peptides and Their Potential Applications.Antibiotics (Basel). 2024 Aug 28;13(9):816. doi: 10.3390/antibiotics13090816. Antibiotics (Basel). 2024. PMID: 39334990 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

