RAR-Dependent and RAR-Independent RXR Signaling in Stem-like Glioma Cells
- PMID: 38003656
- PMCID: PMC10671216
- DOI: 10.3390/ijms242216466
RAR-Dependent and RAR-Independent RXR Signaling in Stem-like Glioma Cells
Abstract
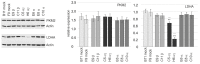
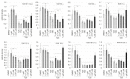
Retinoic acid (RA) exerts pleiotropic effects during neural development and regulates homeostasis in the adult human brain. The RA signal may be transduced through RXR (retinoid-X receptor)-non-permissive RA receptor/RXR heterodimers or through RXR-permissive RXR heterodimers. The significance of RA signaling in malignant brain tumors such as glioblastoma multiforme (GBM) and gliosarcoma (GS) is poorly understood. In particular, the impact RA has on the proliferation, survival, differentiation, or metabolism of GBM- or GS-derived cells with features of stem cells (SLGCs) remains elusive. In the present manuscript, six GBM- and two GS-derived SLGC lines were analyzed for their responsiveness to RAR- and RXR-selective agonists. Inhibition of proliferation and initiation of differentiation were achieved with a RAR-selective pan-agonist in a subgroup of SLGC lines, whereas RXR-selective pan-agonists (rexinoids) supported proliferation in most SLGC lines. To decipher the RAR-dependent and RAR-independent effects of RXR, the genes encoding the RAR or RXR isotypes were functionally inactivated by CRISPR/Cas9-mediated editing in an IDH1-/p53-positive SLGC line with good responsiveness to RA. Stemness, differentiation capacity, and growth behavior were preserved after editing. Taken together, this manuscript provides evidence about the positive impact of RAR-independent RXR signaling on proliferation, survival, and tumor metabolism in SLGCs.
Keywords: CRISPR/Cas9 editing; malignant glioma; retinoic acid receptors; retinoid-X receptor; synthetic retinoids.
Conflict of interest statement
We declare that we have no potential conflict of interest.
Figures
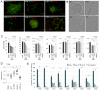
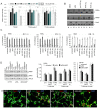
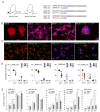


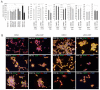

References
-
- Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

