TFIIB-Related Protein BRP5/PTF2 Is Required for Both Male and Female Gametogenesis and for Grain Formation in Rice
- PMID: 38003663
- PMCID: PMC10671200
- DOI: 10.3390/ijms242216473
TFIIB-Related Protein BRP5/PTF2 Is Required for Both Male and Female Gametogenesis and for Grain Formation in Rice
Abstract
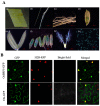
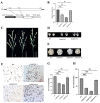
Transcription factor IIB (TFIIB) is a general transcription factor for RNA polymerase II, exerting its influence across various biological contexts. In the majority of eukaryotes, TFIIB typically has two homologs, serving as general transcription factors for RNA polymerase I and III. In plants, however, the TFIIB-related protein family has expanded greatly, with 14 and 9 members in Arabidopsis and rice, respectively. BRP5/pollen-expressed transcription factor 2 (PTF2) proteins belong to a subfamily of TFIIB-related proteins found only in plants and algae. The prior analysis of an Arabidopsis atbrp5 mutant, characterized by a T-DNA insertion at the 5' untranslated region, demonstrated the essential role of BRP5/PTF2 during the process of pollen germination and embryogenesis in Arabidopsis. Using a rice transformation system based on CRISPR/Cas9 technology, we have generated transgenic rice plants containing loss-of-function frameshift mutations in the BRP5/PTF2 gene. Unlike in the Arabidopsis atbrp5 mutant, the brp5/ptf2 frameshift mutations were not transmitted to progeny in rice, indicating an essential role of BRP5/PTF2 in both male and female gamete development or viability. The silencing of rice BRP5/PTF2 expression through RNA interference (RNAi) had little effect on vegetative growth and panicle formation but strongly affected pollen development and grain formation. Genetic analysis revealed that strong RNAi silencing of rice BRP5/PTF2 was still transmissible to progeny almost exclusively through female gametes, as found in the Arabidopsis atbrp5 knockdown mutant. Thus, reduced rice BRP5/PTF2 expression impacted pollen preferentially by interfering with male gamete development or viability. Drawing upon these findings, we posit that BRP5/PTF2 assumes a distinct and imperative function in the realm of plant sexual reproduction.
Keywords: TFIIB; gametogenesis; pollen development; rice; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Pollen-expressed transcription factor 2 encodes a novel plant-specific TFIIB-related protein that is required for pollen germination and embryogenesis in Arabidopsis.Mol Plant. 2013 Jul;6(4):1091-108. doi: 10.1093/mp/sst083. Epub 2013 May 27. Mol Plant. 2013. PMID: 23713077
-
The Arabidopsis transcription factor IIB-related protein BRP4 is involved in the regulation of mitotic cell-cycle progression during male gametogenesis.J Exp Bot. 2014 Jun;65(9):2521-31. doi: 10.1093/jxb/eru140. Epub 2014 Apr 10. J Exp Bot. 2014. PMID: 24723406 Free PMC article.
-
Emergence and expansion of TFIIB-like factors in the plant kingdom.Gene. 2013 Aug 15;526(1):30-8. doi: 10.1016/j.gene.2013.04.022. Epub 2013 Apr 20. Gene. 2013. PMID: 23608173 Free PMC article.
-
Expansion and Functional Diversification of TFIIB-Like Factors in Plants.Int J Mol Sci. 2021 Jan 23;22(3):1078. doi: 10.3390/ijms22031078. Int J Mol Sci. 2021. PMID: 33498602 Free PMC article. Review.
-
Development and genetic regulation of pollen intine in Arabidopsis and rice.Gene. 2024 Jan 30;893:147936. doi: 10.1016/j.gene.2023.147936. Epub 2023 Oct 29. Gene. 2024. PMID: 38381507 Review.
References
-
- Cox M.M., Doudna J., O’Donnell M. Molecular Biology: Principle and Practice. Freenman and Company; New York, NY, USA: 2012.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

