Genome-Wide Identification and Analysis of the ABCF Gene Family in Triticum aestivum
- PMID: 38003668
- PMCID: PMC10671407
- DOI: 10.3390/ijms242216478
Genome-Wide Identification and Analysis of the ABCF Gene Family in Triticum aestivum
Abstract
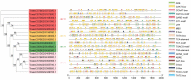
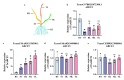
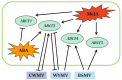
The ATP-binding cassette (ABC) superfamily of proteins is a group of evolutionarily conserved proteins. The ABCF subfamily is involved in ribosomal synthesis, antibiotic resistance, and transcriptional regulation. However, few studies have investigated the role of ABCF in wheat (Triticum aestivum) immunity. Here, we identified 18 TaABCFs and classified them into four categories based on their domain characteristics. Functional similarity between Arabidopsis and wheat ABCF genes was predicted using phylogenetic analysis. A comprehensive genome-wide analysis of gene structure, protein motifs, chromosomal location, and cis-acting elements was also performed. Tissue-specific analysis and expression profiling under temperature, hormonal, and viral stresses were performed using real-time quantitative reverse transcription polymerase chain reaction after randomly selecting one gene from each group. The results revealed that all TaABCF genes had the highest expression at 25 °C and responded to methyl jasmonate induction. Notably, TaABCF2 was highly expressed in all tissues except the roots, and silencing it significantly increased the accumulation of Chinese wheat mosaic virus or wheat yellow mosaic virus in wheat leaves. These results indicated that TaABCF may function in response to viral infection, laying the foundation for further studies on the mechanisms of this protein family in plant defence.
Keywords: ABCF gene family; CWMV; biotic stress; expression; genome-wide; wheat.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








Similar articles
-
Genome-wide identification and expression analysis of the GSK gene family in wheat (Triticum aestivum L.).Mol Biol Rep. 2022 Apr;49(4):2899-2913. doi: 10.1007/s11033-021-07105-2. Epub 2022 Jan 27. Mol Biol Rep. 2022. PMID: 35083611
-
Genome-Wide Identification and Characterization of the Brassinazole-resistant (BZR) Gene Family and Its Expression in the Various Developmental Stage and Stress Conditions in Wheat (Triticum aestivum L.).Int J Mol Sci. 2021 Aug 14;22(16):8743. doi: 10.3390/ijms22168743. Int J Mol Sci. 2021. PMID: 34445448 Free PMC article.
-
Genome-Wide Identification and Characterization of PIN-FORMED (PIN) Gene Family Reveals Role in Developmental and Various Stress Conditions in Triticum aestivum L.Int J Mol Sci. 2021 Jul 9;22(14):7396. doi: 10.3390/ijms22147396. Int J Mol Sci. 2021. PMID: 34299014 Free PMC article.
-
Genome-wide characterization of JASMONATE-ZIM DOMAIN transcription repressors in wheat (Triticum aestivum L.).BMC Genomics. 2017 Feb 13;18(1):152. doi: 10.1186/s12864-017-3582-0. BMC Genomics. 2017. PMID: 28193162 Free PMC article.
-
Genome-wide identification and expression analysis of expansin gene family in common wheat (Triticum aestivum L.).BMC Genomics. 2019 Feb 1;20(1):101. doi: 10.1186/s12864-019-5455-1. BMC Genomics. 2019. PMID: 30709338 Free PMC article.
Cited by
-
Genome-wide identification and characterization of ALKB homolog gene family in wheat (Triticum aestivum L.).Front Plant Sci. 2025 Mar 18;16:1544879. doi: 10.3389/fpls.2025.1544879. eCollection 2025. Front Plant Sci. 2025. PMID: 40171482 Free PMC article.
-
Genome-Wide Identification and Characterization of the Pirin Gene Family in Nicotiana benthamiana.Genes (Basel). 2025 Jan 22;16(2):121. doi: 10.3390/genes16020121. Genes (Basel). 2025. PMID: 40004449 Free PMC article.
References
-
- Raichaudhuri A., Peng M., Naponelli V., Chen S., Sánchez-Fernández R., Gu H., Gregory JF 3rd Hanson A.D., Rea P.A. Plant Vacuolar ATP-binding Cassette Transporters That Translocate Folates and Antifolates in Vitro and Contribute to Antifolate Tolerance in Vivo. J. Biol. Chem. 2009;284:8449–8460. doi: 10.1074/jbc.M808632200. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

