Differential Modulation of Saliva-Derived Microcosm Biofilms by Antimicrobial Peptide LL-31 and D-LL-31
- PMID: 38003760
- PMCID: PMC10675243
- DOI: 10.3390/pathogens12111295
Differential Modulation of Saliva-Derived Microcosm Biofilms by Antimicrobial Peptide LL-31 and D-LL-31
Abstract
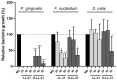
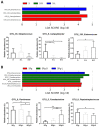
Microbiome modulation, aiming to restore a health-compatible microbiota, is a novel strategy to treat periodontitis. This study evaluated the modulation effects of antimicrobial peptide LL-31 and its D-enantiomer (D-LL-31) on saliva-derived microcosm biofilms, spiked with or without Porphyromonas gingivalis. To this end, one-day-old biofilms were incubated for 24 h with biofilm medium alone, or medium containing 40 µM LL-31 or D-LL-31, after which biofilms were grown for 5 days. Biofilms were assessed at 1 day and 5 days after intervention for the total viable cell counts, dipeptidyl peptidase IV (DPP4) activity, P. gingivalis amount (by qPCR) and microbial composition (by sequencing). The results showed that D-LL-31, not LL-31, significantly reduced the total viable cell counts, the P. gingivalis amount, and the DPP4 activity of the biofilms spiked with P. gingivalis, but only at 1 day after intervention. In the biofilms spiked with P. gingivalis, D-LL-31 tended to reduce the α-diversity and the compositional shift of the biofilms in time as compared to the control and LL-31 groups. In conclusion, D-LL-31 showed a better performance than LL-31 in biofilm modulation. The biofilm modulation function of the peptides could be impaired when the biofilms were in a severely dysbiotic state.
Keywords: 16S rRNA gene amplicon sequencing; Porphyromonas gingivalis; oral microbiome; periodontitis; saliva-derived microcosm biofilms.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Manipulation of Saliva-Derived Microcosm Biofilms To Resemble Dysbiotic Subgingival Microbiota.Appl Environ Microbiol. 2021 Jan 15;87(3):e02371-20. doi: 10.1128/AEM.02371-20. Print 2021 Jan 15. Appl Environ Microbiol. 2021. PMID: 33158898 Free PMC article.
-
Phellodendron bark extract and berberine chloride suppress microbiome dysbiosis in a saliva-derived in vitro microcosm biofilm model.Arch Oral Biol. 2025 Jun;174:106231. doi: 10.1016/j.archoralbio.2025.106231. Epub 2025 Mar 22. Arch Oral Biol. 2025. PMID: 40209653
-
Regrowth of Microcosm Biofilms on Titanium Surfaces After Various Antimicrobial Treatments.Front Microbiol. 2019 Nov 25;10:2693. doi: 10.3389/fmicb.2019.02693. eCollection 2019. Front Microbiol. 2019. PMID: 31824464 Free PMC article.
-
Microcosm biofilms cultured from different oral niches in periodontitis patients.J Oral Microbiol. 2018 Nov 27;11(1):1551596. doi: 10.1080/20022727.2018.1551596. eCollection 2019. J Oral Microbiol. 2018. PMID: 30598734 Free PMC article.
-
An in vitro model demonstrating homeostatic interactions between reconstructed human gingiva and a saliva-derived multispecies biofilm.Microbiome. 2025 Feb 28;13(1):58. doi: 10.1186/s40168-025-02033-w. Microbiome. 2025. PMID: 40022258 Free PMC article.
Cited by
-
The oral-gut microbiome axis in inflammatory bowel disease: from inside to insight.Front Immunol. 2024 Jul 26;15:1430001. doi: 10.3389/fimmu.2024.1430001. eCollection 2024. Front Immunol. 2024. PMID: 39131163 Free PMC article. Review.
-
Advancing Nanotechnology: Targeting Biofilm-Forming Bacteria with Antimicrobial Peptides.BME Front. 2025 Mar 4;6:0104. doi: 10.34133/bmef.0104. eCollection 2025. BME Front. 2025. PMID: 40041091 Free PMC article. Review.
-
Natural Antimicrobial Peptides and Their Synthetic Analogues for Effective Oral Microflora Control and Oral Infection Treatment-The Role of Ceragenins in the Development of New Therapeutic Methods.Pharmaceuticals (Basel). 2024 Dec 20;17(12):1725. doi: 10.3390/ph17121725. Pharmaceuticals (Basel). 2024. PMID: 39770567 Free PMC article. Review.
-
The Oral Microbiome and Us.Adv Exp Med Biol. 2025;1472:3-9. doi: 10.1007/978-3-031-79146-8_1. Adv Exp Med Biol. 2025. PMID: 40111682 Review.
References
-
- Alassy H., Pizarek J.A., Kormas I., Pedercini A., Wolff L.F. Antimicrobial adjuncts in the management of periodontal and peri-implant diseases and conditions: A narrative review. Front. Oral. Maxillofac. Med. 2021;3:16. doi: 10.21037/fomm-20-84. - DOI
-
- Ramanauskaite E., Moraschini V., Machiulskiene V., Sculean A. Clinical efficacy of single and multiple applications of antimicrobial photodynamic therapy in periodontal maintenance: A systematic review and network meta-analysis. Photodiagnosis Photodyn. Ther. 2021;36:102435. doi: 10.1016/j.pdpdt.2021.102435. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

