Implications of Flagellar Attachment Zone Proteins TcGP72 and TcFLA-1BP in Morphology, Proliferation, and Intracellular Dynamics in Trypanosoma cruzi
- PMID: 38003831
- PMCID: PMC10675206
- DOI: 10.3390/pathogens12111367
Implications of Flagellar Attachment Zone Proteins TcGP72 and TcFLA-1BP in Morphology, Proliferation, and Intracellular Dynamics in Trypanosoma cruzi
Abstract
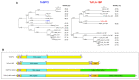
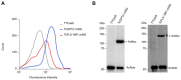
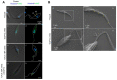
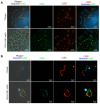
The highly adaptable parasite Trypanosoma cruzi undergoes complex developmental stages to exploit host organisms effectively. Each stage involves the expression of specific proteins and precise intracellular structural organization. These morphological changes depend on key structures that control intracellular components' growth and redistribution. In trypanosomatids, the flagellar attachment zone (FAZ) connects the flagellum to the cell body and plays a pivotal role in cell expansion and structural rearrangement. While FAZ proteins are well-studied in other trypanosomatids, there is limited knowledge about specific components, organization, and function in T. cruzi. This study employed the CRISPR/Cas9 system to label endogenous genes and conduct deletions to characterize FAZ-specific proteins during epimastigote cell division and metacyclogenesis. In T. cruzi, these proteins exhibited distinct organization compared to their counterparts in T. brucei. TcGP72 is anchored to the flagellar membrane, while TcFLA-1BP is anchored to the membrane lining the cell body. We identified unique features in the organization and function of the FAZ in T. cruzi compared to other trypanosomatids. Deleting these proteins had varying effects on intracellular structures, cytokinesis, and metacyclogenesis. This study reveals specific variations that directly impact the success of cell division and differentiation of this parasite.
Keywords: CRISPR/Cas9; flagellar attachment zone; morphological changes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Flagellar pocket restructuring through the Leishmania life cycle involves a discrete flagellum attachment zone.J Cell Sci. 2016 Feb 15;129(4):854-67. doi: 10.1242/jcs.183152. Epub 2016 Jan 8. J Cell Sci. 2016. PMID: 26746239 Free PMC article.
-
The flagellar attachment zone of Trypanosoma cruzi epimastigote forms.J Struct Biol. 2006 Apr;154(1):89-99. doi: 10.1016/j.jsb.2005.11.008. Epub 2005 Dec 22. J Struct Biol. 2006. PMID: 16414276
-
CRISPR/Cas9-Induced Disruption of Paraflagellar Rod Protein 1 and 2 Genes in Trypanosoma cruzi Reveals Their Role in Flagellar Attachment.mBio. 2015 Jul 21;6(4):e01012. doi: 10.1128/mBio.01012-15. mBio. 2015. PMID: 26199333 Free PMC article.
-
Basic Biology of Trypanosoma cruzi.Curr Pharm Des. 2021;27(14):1671-1732. doi: 10.2174/1381612826999201203213527. Curr Pharm Des. 2021. PMID: 33272165 Review.
-
The Flagellum Attachment Zone: 'The Cellular Ruler' of Trypanosome Morphology.Trends Parasitol. 2016 Apr;32(4):309-324. doi: 10.1016/j.pt.2015.12.010. Epub 2016 Jan 8. Trends Parasitol. 2016. PMID: 26776656 Free PMC article. Review.
Cited by
-
Kharon Is Crucial for Trypanosoma cruzi Morphology but Does Not Impair In Vitro Infection.Pathogens. 2025 Mar 25;14(4):312. doi: 10.3390/pathogens14040312. Pathogens. 2025. PMID: 40333073 Free PMC article.
-
Contribution of microscopy to a better understanding of the anatomy of pathogenic protists.Proc Natl Acad Sci U S A. 2024 Apr 23;121(17):e2321515121. doi: 10.1073/pnas.2321515121. Epub 2024 Apr 15. Proc Natl Acad Sci U S A. 2024. PMID: 38621128 Free PMC article.
References
-
- De Souza W. International Review of Cytology. Volume 86. Academic Press; Cambridge, MA, USA: 1984. Cell Biology of Trypanosoma cruzi; pp. 197–283. - PubMed
LinkOut - more resources
Full Text Sources

