Network Pharmacology, Molecular Docking, and Molecular Dynamics Simulation to Elucidate the Molecular Targets and Potential Mechanism of Phoenix dactylifera (Ajwa Dates) against Candidiasis
- PMID: 38003833
- PMCID: PMC10674288
- DOI: 10.3390/pathogens12111369
Network Pharmacology, Molecular Docking, and Molecular Dynamics Simulation to Elucidate the Molecular Targets and Potential Mechanism of Phoenix dactylifera (Ajwa Dates) against Candidiasis
Abstract
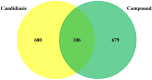
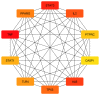
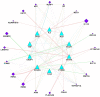
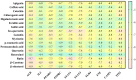
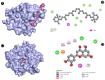
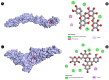
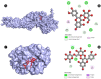
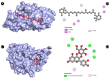
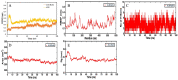
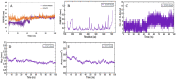
Candidiasis, caused by opportunistic fungal pathogens of the Candida genus, poses a significant threat to immunocompromised individuals. Natural compounds derived from medicinal plants have gained attention as potential sources of anti-fungal agents. Ajwa dates (Phoenix dactylifera L.) have been recognized for their diverse phytochemical composition and therapeutic potential. In this study, we employed a multi-faceted approach to explore the anti-candidiasis potential of Ajwa dates' phytochemicals. Utilizing network pharmacology, we constructed an interaction network to elucidate the intricate relationships between Ajwa dates phytoconstituents and the Candida-associated molecular targets of humans. Our analysis revealed key nodes in the network (STAT3, IL-2, PTPRC, STAT1, CASP1, ALB, TP53, TLR4, TNF and PPARG), suggesting the potential modulation of several crucial processes (the regulation of the response to a cytokine stimulus, regulation of the inflammatory response, positive regulation of cytokine production, cellular response to external stimulus, etc.) and fungal pathways (Th17 cell differentiation, the Toll-like receptor signaling pathway, the C-type lectin receptor signaling pathway and necroptosis). To validate these findings, molecular docking studies were conducted, revealing the binding affinities of the phytochemicals towards selected Candida protein targets of humans (ALB-rutin (-9.7 kJ/mol), STAT1-rutin (-9.2 kJ/mol), STAT3-isoquercetin (-8.7 kJ/mol), IL2-β-carotene (-8.5 kJ/mol), CASP1-β-carotene (-8.2 kJ/mol), TP53-isoquercetin (-8.8 kJ/mol), PPARG-luteolin (-8.3 kJ/mol), TNF-βcarotene (-7.7 kJ/mol), TLR4-rutin (-7.4 kJ/mol) and PTPRC-rutin (-7.0 kJ/mol)). Furthermore, molecular dynamics simulations of rutin-ALB and rutin-STAT1 complex were performed to gain insights into the stability and dynamics of the identified ligand-target complexes over time. Overall, the results not only contribute to the understanding of the molecular interactions underlying the anti-fungal potential of specific phytochemicals of Ajwa dates in humans but also provide a rational basis for the development of novel therapeutic strategies against candidiasis in humans. This study underscores the significance of network pharmacology, molecular docking and dynamics simulations in accelerating the discovery of natural products as effective anti-fungal agents. However, further experimental validation of the identified compounds is warranted to translate these findings into practical therapeutic applications.
Keywords: Ajwa dates; Phoenix dactylifera; candidiasis; fungal infection; molecular dynamics; network pharmacology.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


Similar articles
-
Network Pharmacology and In silico Elucidation of Phytochemicals Extracted from Ajwa Dates (Phoenix dactylifera L.) to Inhibit Akt and PI3K Causing Triple Negative Breast Cancer (TNBC).Curr Pharm Des. 2025;31(10):774-796. doi: 10.2174/0113816128348876241017101729. Curr Pharm Des. 2025. PMID: 39698883
-
Ajwa date extract (Phoenix dactylifera L.): Phytochemical analysis, antiviral activity against herpes simplex virus-I and coxsackie B4 virus, and in silico study.Saudi Med J. 2025 Jan;46(1):26-35. doi: 10.15537/smj.2025.46.1.20240780. Saudi Med J. 2025. PMID: 39779363 Free PMC article.
-
Anti-inflammatory and antibacterial potential of Ajwa date (Phoenix dactylifera L.) extract in burn infection.J Adv Pharm Technol Res. 2023 Jul-Sep;14(3):161-165. doi: 10.4103/japtr.japtr_138_23. Epub 2023 Jul 28. J Adv Pharm Technol Res. 2023. PMID: 37692010 Free PMC article. Review.
-
Exploring shared therapeutic targets for Alzheimer's disease and glioblastoma using network pharmacology and protein-protein interaction approach.Front Chem. 2025 Mar 12;13:1549186. doi: 10.3389/fchem.2025.1549186. eCollection 2025. Front Chem. 2025. PMID: 40144222 Free PMC article.
-
Date Palm (Phoenix dactylifera): Novel Findings and Future Directions for Food and Drug Discovery.Curr Drug Discov Technol. 2019;16(1):2-10. doi: 10.2174/1570163815666180320111937. Curr Drug Discov Technol. 2019. PMID: 29557751 Review.
Cited by
-
Unraveling the Core Components and Critical Targets of Houttuynia cordata Thunb. in Treating Non-small Cell Lung Cancer through Network Pharmacology and Multi-omics Analysis.Curr Pharm Des. 2025;31(7):540-558. doi: 10.2174/0113816128330427241017110325. Curr Pharm Des. 2025. PMID: 39440769 Free PMC article.
-
Quercetin Exhibits Broad-Spectrum Antibiofilm and Antiquorum Sensing Activities Against Gram-Negative Bacteria: In Vitro and In Silico Investigation Targeting Antimicrobial Therapy.Can J Infect Dis Med Microbiol. 2025 Mar 30;2025:2333207. doi: 10.1155/cjid/2333207. eCollection 2025. Can J Infect Dis Med Microbiol. 2025. PMID: 40196379 Free PMC article.
-
Mechanistic Insights into the Anticancer Potential of Asparagus racemosus Willd. Against Triple-Negative Breast Cancer: A Network Pharmacology and Experimental Validation Study.Pharmaceuticals (Basel). 2025 Mar 19;18(3):433. doi: 10.3390/ph18030433. Pharmaceuticals (Basel). 2025. PMID: 40143209 Free PMC article.
-
Insights into the Therapeutic Targets and Molecular Mechanisms of Eruca sativa Against Colorectal Cancer: An Integrated Approach Combining Network Pharmacology, Molecular Docking and Dynamics Simulation.Pharmaceuticals (Basel). 2025 Mar 24;18(4):453. doi: 10.3390/ph18040453. Pharmaceuticals (Basel). 2025. PMID: 40283891 Free PMC article.
-
Integrative approach to decipher pharmacological mechanism of Cinnamomum zeylanicum essential oil in prostate cancer.Med Oncol. 2025 Mar 12;42(4):100. doi: 10.1007/s12032-025-02665-w. Med Oncol. 2025. PMID: 40072751
References
-
- Saini S. Candidiasis: Past, present and future. Int. J. Infect. Trop. Dis. 2015;2:12–24.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

