Transcriptional Profiling of SARS-CoV-2-Infected Calu-3 Cells Reveals Immune-Related Signaling Pathways
- PMID: 38003837
- PMCID: PMC10674242
- DOI: 10.3390/pathogens12111373
Transcriptional Profiling of SARS-CoV-2-Infected Calu-3 Cells Reveals Immune-Related Signaling Pathways
Abstract
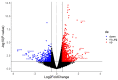
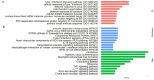
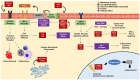
The COVID-19 disease, caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), emerged in late 2019 and rapidly spread worldwide, becoming a pandemic that infected millions of people and caused significant deaths. COVID-19 continues to be a major threat, and there is a need to deepen our understanding of the virus and its mechanisms of infection. To study the cellular responses to SARS-CoV-2 infection, we performed an RNA sequencing of infected vs. uninfected Calu-3 cells. Total RNA was extracted from infected (0.5 MOI) and control Calu-3 cells and converted to cDNA. Sequencing was performed, and the obtained reads were quality-analyzed and pre-processed. Differential expression was assessed with the EdgeR package, and functional enrichment was performed in EnrichR for Gene Ontology, KEGG pathways, and WikiPathways. A total of 1040 differentially expressed genes were found in infected vs. uninfected Calu-3 cells, of which 695 were up-regulated and 345 were down-regulated. Functional enrichment analyses revealed the predominant up-regulation of genes related to innate immune response, response to virus, inflammation, cell proliferation, and apoptosis. These transcriptional changes following SARS-CoV-2 infection may reflect a cellular response to the infection and help to elucidate COVID-19 pathogenesis, in addition to revealing potential biomarkers and drug targets.
Keywords: COVID-19; Calu-3 cells; RNA-seq; host-pathogen interaction; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Long-Read RNA Sequencing Identifies Polyadenylation Elongation and Differential Transcript Usage of Host Transcripts During SARS-CoV-2 In Vitro Infection.Front Immunol. 2022 Apr 6;13:832223. doi: 10.3389/fimmu.2022.832223. eCollection 2022. Front Immunol. 2022. PMID: 35464437 Free PMC article.
-
Novel signaling pathways regulate SARS-CoV and SARS-CoV-2 infectious disease.Medicine (Baltimore). 2021 Feb 19;100(7):e24321. doi: 10.1097/MD.0000000000024321. Medicine (Baltimore). 2021. PMID: 33607766 Free PMC article.
-
Protein Coding and Long Noncoding RNA (lncRNA) Transcriptional Landscape in SARS-CoV-2 Infected Bronchial Epithelial Cells Highlight a Role for Interferon and Inflammatory Response.Genes (Basel). 2020 Jul 7;11(7):760. doi: 10.3390/genes11070760. Genes (Basel). 2020. PMID: 32646047 Free PMC article.
-
Ribosome-Profiling Reveals Restricted Post Transcriptional Expression of Antiviral Cytokines and Transcription Factors during SARS-CoV-2 Infection.Int J Mol Sci. 2021 Mar 25;22(7):3392. doi: 10.3390/ijms22073392. Int J Mol Sci. 2021. PMID: 33806254 Free PMC article.
-
Highlight of Immune Pathogenic Response and Hematopathologic Effect in SARS-CoV, MERS-CoV, and SARS-Cov-2 Infection.Front Immunol. 2020 May 12;11:1022. doi: 10.3389/fimmu.2020.01022. eCollection 2020. Front Immunol. 2020. PMID: 32574260 Free PMC article. Review.
Cited by
-
Discovery of a pan anti-SARS-CoV-2 monoclonal antibody with highly efficient infected cell killing capacity for novel immunotherapeutic approaches.Emerg Microbes Infect. 2025 Dec;14(1):2432345. doi: 10.1080/22221751.2024.2432345. Epub 2024 Dec 9. Emerg Microbes Infect. 2025. PMID: 39584380 Free PMC article.
-
Inhibition of complement system-related gene ITGB2 attenuates epithelial-mesenchymal transition and inflammation in diabetic nephropathy.Eur J Med Res. 2025 Feb 8;30(1):87. doi: 10.1186/s40001-025-02323-x. Eur J Med Res. 2025. PMID: 39920798 Free PMC article.
-
Identification of Factors Associated with Mortality in the Elderly Population with SARS-CoV-2 Infection: Results from a Longitudinal Observational Study from Romania.Pharmaceuticals (Basel). 2024 Feb 3;17(2):202. doi: 10.3390/ph17020202. Pharmaceuticals (Basel). 2024. PMID: 38399417 Free PMC article.
-
Optimizing Neutrophil Recruitment to Tackle Bacterial Infections.Am J Respir Cell Mol Biol. 2025 Jul;73(1):3-5. doi: 10.1165/rcmb.2024-0575ED. Am J Respir Cell Mol Biol. 2025. PMID: 39836037 Free PMC article. No abstract available.
References
-
- World Health Organization COVID-19 Weekly Epidemiological Update. 2023. [(accessed on 24 August 2023)]. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on....
-
- Aiewsakun P., Phumiphanjarphak W., Ludowyke N., Purwono P.B., Manopwisedjaroen S., Srisaowakarn C., Ekronarongchai S., Suksatu A., Yuvaniyama J., Thitithanyanont A. Systematic Exploration of SARS-CoV-2 Adaptation to Vero E6, Vero E6/TMPRSS2, and Calu-3 Cells. Genome Biol. Evol. 2023;15:evad035. doi: 10.1093/gbe/evad035. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

